Talk:Knight:Beta-galactosidase assay/96 well format
From OpenWetWare
Jump to navigationJump to search
This is an outline of various control experiments that I need to do. It is a work in progress and has not been done.
A600 versus cell density
- Grow an overnight culture to saturation in EZ Rich Media
- Pellet the cells
- Resuspend in 1/4 of the original volume with EZ Rich Media
- Add 350 μL of cell suspension to the first well
- Add 175 μL of previous well to next well.
- Add 175 μL of EZ Rich Media to that well.
- Repeat dilution series until the well's solution looks totally clear.
- Add an additional well of 175 μL EZ Rich Media.
- Measure A600 of each well in the plate reader.
- Plot A420 versus dilution factor. This relationship should be linear.
Effect of evaporation on absorbance readings
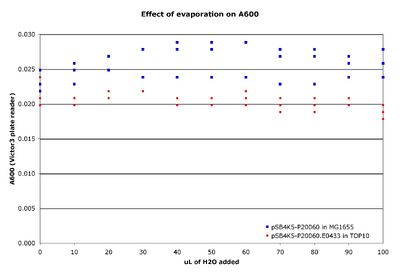
Effect of evaporation on A600 readings
- Aliquot 50 μL of culture into an entire row of wells.
- Do duplicates rows to assess measurement variability in duplicate samples.
- Aliquot 100 μL of a of culture into a second row of wells.
- Add increasing volumes of water to each well (by 10 μL increments).
- Measure the A600 of the plate.
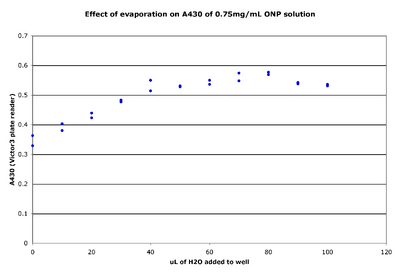
Effect of evaporation on A420 readings
- Aliquot 50 μL of a set concentration of ONP into an entire row of wells.
- Could add different concentrations of ONP to different rows.
- Add increasing volumes of water to each well (by 10 μL increments).
- Measure the A420 of the plate.
β-galactosidase activity versus number of cells
- Do parallel β-galactosidase assays with a variable volumes of cells from the same grown culture.
- 1 = 1 μL of cells
- 2 = 2 μL of cells
- 3 = 3 μL of cells
- 4 = 4 μL of cells
- 5 = 5 μL of cells
- 6 = 5 μL of media
β-galactosidase activity versus growth phase of culture
- Grow an overnight culture of several constructs
- A = P20060 in MG1655
- B = P20060 in MG1655+IPTG
- C = P20060+IPTG+AHL
- D = P20060.E0433+IPTG
- E = P20060.E0433+IPTG+AHL
- F = R2000.E0433+IPTG+AHL
- G =
- H = Media+IPTG+AHL
- In the morning, dilute back samples into EZ Rich Media to an A600 of 0.001 (via a Nanodrop reading) in tubes (5 mL of culture in a 14mL tubes per construct).
- Let grow 1 hour.
- Add IPTG and/or AHL to cultures as appropriate.
- Prepare 8mL of permeabilization buffer.
- Every hour
- Take 175 μL of each culture and put it in the appropriate row and column of a 96 well plate (A600 plate).
- Measure the absorbance at 600nm in the plate reader of the newest column of the A600 plate.
- Aliquot 80 μL permeabilization buffer into each well of the corresponding column of a second 96 well plate (permeabilization plate).
- Take 20 μL of culture from the most recent column of the A600 plate and add it to the corresponding column of the permeabilization plate.
- Parafilm that column of the permeabilization plate.
- Once each time point has been taken, move 25 μL of each well from the permeabilized culture to a new plate.
- Add 150 μL of substrate solution to each well.
- Place plate in the plate reader to measure change in A430 as a function of time.
- Plot the β-galactosidase activity in Miller Units as a function of the A600 of the culture.
A420 versus o-nitrophenol concentration
- Make up a solution of 14 mL to control for background absorbance in β-galactosidase assays
- 1600 μL permeabilization solution
- 12 mL substrate solution without ONPG
- 400 μL EZ rich media supplemented with kanamycin and AHL
- Make up 1mL of 1 mg/mL ONP.
- Add 350 μL of 1mg/mL solution to the first well.
- Move 175 μL of previous well to next well.
- Add 175 μL of background solution to that well.
- Repeat dilution series until the well's solution looks totally clear.
- Add an additional well of 175 μL background solution.
- Measure A420 of each well in the plate reader.
- Plot A420 versus ONP concentration. This relationship should be linear.
- Reshma 13:16, 13 November 2007 (CST): I seem to get different calibration curves every time I do this experiment presumably due to errors in measurement of the ONP (since I am measuring small amounts).