BME103:T930 Group 16 l2: Difference between revisions
| Line 105: | Line 105: | ||
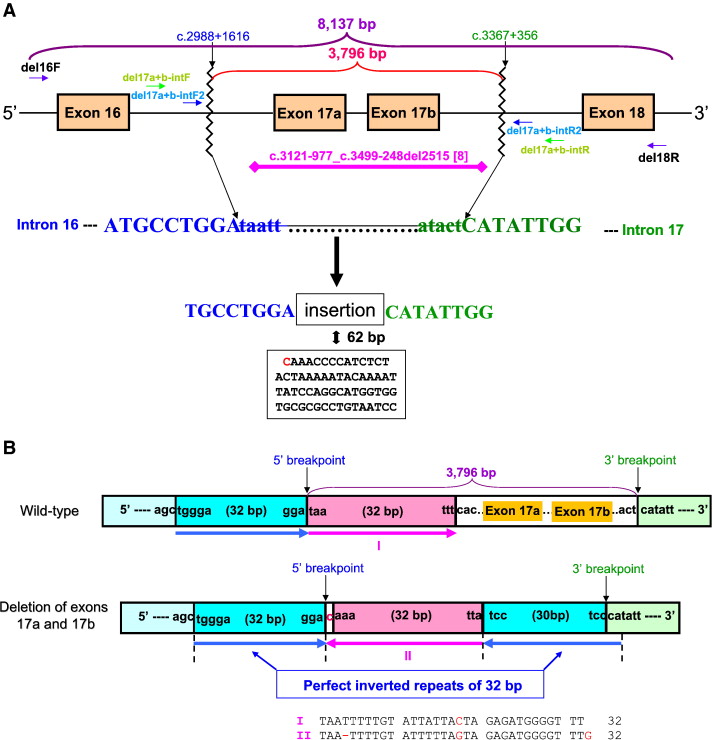
[[Image:Cystic Fibrosis.jpg]]<br> | [[Image:Cystic Fibrosis.jpg]]<br> | ||
Large deletions in cystic fibrosis alleles have been estimated to constitute 1–2% pathogenic alleles, but the occurrence could be much higher in classical cystic fibrosis patients with one mutation detectable by the routine screening/sequencing work-up. The rearranged region is flanked by a pair of perfectly inverted repeats of 32 bp. The start and end of the 32 bp repeats (blue arrows) are denoted by vertical dashed lines. The nucleotide sequence alignment shows that I and II (reverse-complemented) are highly homologous. | Large deletions in cystic fibrosis alleles have been estimated to constitute 1–2% pathogenic alleles, but the occurrence could be much higher in classical cystic fibrosis patients with one mutation detectable by the routine screening/sequencing work-up. The rearranged region is flanked by a pair of perfectly inverted repeats of 32 bp. The start and end of the 32 bp repeats (blue arrows) are denoted by vertical dashed lines. The nucleotide sequence alignment shows that I and II (reverse-complemented) are highly homologous. | ||
http://www.sciencedirect.com/science/article/pii/S1569199312001737 | *http://www.sciencedirect.com/science/article/pii/S1569199312001737 | ||
http://www.bmb.msu.edu/faculty/hong.html | *http://www.bmb.msu.edu/faculty/hong.html | ||
<!-- ##### DO NOT edit below this line unless you know what you are doing. ##### --> | <!-- ##### DO NOT edit below this line unless you know what you are doing. ##### --> | ||
|} | |} | ||
Revision as of 19:16, 25 November 2012
| Home People Lab Write-Up 1 Lab Write-Up 2 Lab Write-Up 3 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
OUR TEAM
LAB 2 WRITE-UPThermal Cycler EngineeringOur re-design is based upon the Open PCR system originally designed by Josh Perfetto and Tito Jankowski.
ProtocolsMaterials
PCR Protocol
DNA Measurement Protocol Research and DevelopmentBackground on Disease Markers Group 16 decided to investigate cystic fibrosis, a disease that is caused by mutations in a specific gene on the seventh chromosome, and is then passed down through families as a recessive trait. In this disease, mucus accumulates inside the lungs, digestive tract, and other cavities in the body. This life-threatening disease is the most common chronic lung disease to affect children and is often diagnosable by the age of two. However, weaker strains can go undetected until early adulthood. The effects of this disease, however, have been mediated through the use of treatments that can postpone some of the changes that occur in the lungs. Half of the patients with cystic fibrosis live past 28 years, and patients who only have mucus buildup in the digestive tract are even better off. Gene therapy holds great promise for treating cystic fibrosis. The marker associated with cystic fibrosis is a two nucleotide deletion and has identity rs200007348. This two nucleotide deletion is due to an adenine-guanine swap that occurs in the codon TGG. A codon is a genetic code involved in the RNA process that impacts protein translation. When the TGG undergoes the adenine guanine swap, it becomes TGA which codes for the stop codon, ending protein translation. This SNP heightens susceptibility to cystic fibrosis, a disease with the frequency 1 out of 2000 in Europe and 1 out of 3500 in the United States
Primer Design Reverse primer: 3' CGTCTCTTACTCTATCTCTC 5' Forward primer: 5' AAATATCTGGCTGAGTGTTT 3' Cystic fibrosis is caused by a 3 bp deletion that leads to a protein which lacks a critical phenylalanine amino acid in the protein. PCR primers have been developed that can distinguish a normal gene from a mutant gene. With these primers a 154 bp product is produced from a normal individual and a 151 bp product is amplified from DNA of an individual with the disease. The disease allele is complementary which will result in a positive result in Open PCR. However, a regular allele cannot give a positive result because the lost nucleotides will be added back into the primer causing a frameshift mutation of three. Thus, these primers are built 151 bp apart and this shortens the temperature cycles from 30 seconds to 10 seconds.
Illustration
| ||||||