BioBuilding: Synthetic Biology for Teachers: Lab 1: Difference between revisions
No edit summary |
James Dixon (talk | contribs) No edit summary |
||
| Line 5: | Line 5: | ||
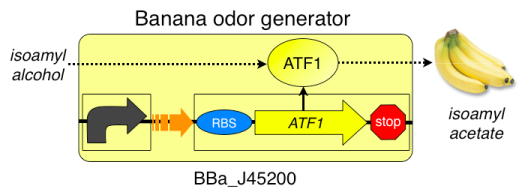
[[Image:Copy of BBa J45200.png]] | [[Image:Copy of BBa J45200.png]] | ||
== Teacher Considerations == | == Teacher Considerations == | ||
This lab provides a valuable opportunity to teach microbiology techniques, population growth dynamics, molecular genetics and basic synthetic biology concepts in a meaningful, real world way. As can be seen in the discussion questions for the lab report, the analysis of the lab will provide the students with a chance to do meaningful error analysis and examine the difference between quantitative results and qualitative results. If time allows the students can carry out all parts of the procedure. This will take about 5 class periods in a typical High School AP Biology or Biotechnology class. If instead the teacher prepares in advance the samples as outlined in Part 1 of the procedure, the students can conduct the smell tests and population measurements over two or three days. In a college lab course, most of the bacterial growth curve can be conducted in a typical three to four hour lab period. The procedure includes instructions for using a spectrophotometer to measure the population growth. If a spectrophotometer is not available, the population can be easily measured using the McFarland Turbidity methodology, as explained below. | This lab provides a valuable opportunity to teach microbiology techniques, population growth dynamics, molecular genetics and basic synthetic biology concepts in a meaningful, real world way. As can be seen in the discussion questions for the lab report, the analysis of the lab will provide the students with a chance to do meaningful error analysis and examine the difference between quantitative results and qualitative results. | ||
This lab offers two different protocols based on the time the teacher wishes to allow. Each of these protocols covers the same concepts but allows for different emphases. | |||
Protocol A: This is a shorter procedure for the students. This shorter protocol allows for more emphasis on microbiological techniques. This protocol is recommended should the teacher choose to have the students do the initial bacterial culturing. Essentially, a day prior to any data collection, the large cultures are set up. Instead of letting them all run, part of the culture is immediately removed and placed in the refrigerator. This will be the lag phase sample. After 5-7 hours, a second sample is removed. This will be the log phase sample. The last third of the culture will be allowed to grow overnight. This will be the stationary phase sample. This will allow the students to easily obtain data in one lab period. 1.3[[Category:1.3]] | |||
If time allows the students can carry out all parts of the procedure. This will take about 5 class periods in a typical High School AP Biology or Biotechnology class. If instead the teacher prepares in advance the samples as outlined in Part 1 of the procedure, the students can conduct the smell tests and population measurements over two or three days. In a college lab course, most of the bacterial growth curve can be conducted in a typical three to four hour lab period. The procedure includes instructions for using a spectrophotometer to measure the population growth. If a spectrophotometer is not available, the population can be easily measured using the McFarland Turbidity methodology, as explained below. | |||
==Needed Materials== | ==Needed Materials== | ||
| Line 39: | Line 44: | ||
* 1 ml isoamyl alcohol | * 1 ml isoamyl alcohol | ||
<br style="clear:both" /> | <br style="clear:both" /> | ||
==Workflow== | ==Workflow Protocol A== | ||
===Classroom Content=== | |||
[[Image:Intro lab1 BioPrimer1.png|thumb|600 px| [http://openwetware.org/images/3/30/BioPrimerNo1.pdf BioPrimer #1 pdf] ]] | |||
*[http://biobuilder.org/ BioBuilder] material that sets up this lesson starts [[Media:BioPrimerNo1.pdf| here]] | |||
*'''Day 1:''' streak strains from stabs onto plates | |||
*'''Day 2:''' grow strains from plates as liquid overnights | |||
*'''Day 3:''' subculture bacteria in larger volumes, place lag phase samples in refrigerator, incubate log phase samples for 5-7 hours and refrigerate, allow stationary phase samples to incubate overnight | |||
*'''Day 4:''' provide students with lag, log and stationary samples for data collection | |||
*When you are done with this lab, [http://www.surveymonkey.com/s/ZP537Z3 here] is a link to survey that you can offer the students. Thank you for helping us improve this content. | |||
===Annotated Procedure=== | |||
===Day 1: === | |||
====TO DO==== | |||
*Streak out strains from stabs to plates<br> | |||
*Prepare banana extract standards | |||
=====Streak out strains from stabs to plates===== | |||
We will be receiving our bacteria with the plasmid already inserted. This culture will come in the form of a "stab" or "slant", a test tube with a small amount of bacteria on a slanted media. To continue the experiment we will have to further culture the bacteria by streaking out the stabs onto LB+amp plates. The plates will be incubated 37° overnight. <br> | |||
# Using a sterile toothpick or inoculating loop, gather a small amount of bacteria from the stab and transfer it to a petri dish containing Luria Broth (LB) agar plus ampicillin medium.<br> | |||
# Repeat with the remaining stab samples, streaking out each onto a different petri dish.<br> | |||
# Place these cultures in a 37°C incubator overnight.<br> | |||
This video illustrates the technique used [http://www.youtube.com/watch?v=QydH5ZoD_Aw for this transfer.]<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''You may wish to conduct this procedure yourself. However, if time allows, the students will enjoy learning these microbiological techniques. If your class will test the whole set, there will be 4 strains to streak out. Strains can also be streaked out on LB+Amp+Cam if you'd like to verify the indole- strain background.</font color> <br> | |||
=====Prepare banana extract standards===== | |||
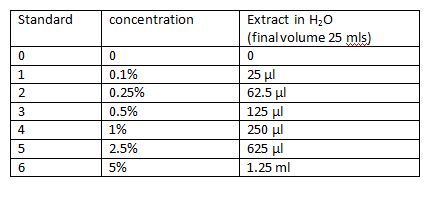
The banana extract is provided in the kit. It will be necessary to make up the standards following the table. [[Image:Banana_extract_formula.JPG]]<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''The banana extract is an oil and will not dissolve in water. However, the concentrations are low and as long as the standard is given a shake before smelling, a suspension is sufficient.</font color> | |||
===Day 2:=== | |||
====TO DO:==== | |||
*Grow liquid overnights of bacterial strains<br> | |||
*Prepare Turbidity standards (if no spectrophotometer is available) | |||
=====Grow liquid overnights of bacterial strains===== | |||
1. Using a sterile inoculating loop, transfer a bacterial colony from one of the petri dishes to a large sterile culture tube containing 5 ml of Luria Broth and 5 μl of ampicillin.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''Colonies can be inoculated into the media with a toothpick, a loop, or a pipet tip.''</font><br> | |||
2. Repeat for each strain you will inoculate.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''Each group will need 2 mls of each sample for the next day of this procedure. If you conducting this part of the procedure for your students, you'll have to scale up the amounts in order to prepare enough for each group.</font><br> | |||
3. Place the culture tubes in the roller wheel in the incubator at 37°C overnight. Be sure to balance the tubes across from each other to minimize stress on the roller wheel.<br> | |||
This video illustrates the general technique for setting up [http://www.youtube.com/watch?v=0odxJy0nR9s&NR=1 overnight liquid cultures], though you’ll be transferring cells from the petri dish to the Luria Broth.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''If you do not have a roller wheel and an incubator, you can prepare these cultures in small erlenmeyer flasks (with stir bars) placed on a stir plate at a slow pace. Cultures are stable and active for a week at least (stored at room temp or in the fridge) but will take considerably longer to start growing on the day you subculture (~3 hours rather than 1).</font><br> | |||
=====Prepare turbidity standards===== | |||
As the populations of bacteria increase, the culture media gets increasingly turbid. Using the [http://en.wikipedia.org/wiki/McFarland_standards McFarland Turbidity Scale], it is possible to estimate the changes in turbidity. The results will not be as precise as what you would measure with a spectrophotometer, but the changes over time will be detected and the results can be graphed. | |||
[[Image:McFarland_table.PNG|center]]<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''these standards can be prepared well in advance of lab and are useful if you are running the protocols without access to a spectrophotometer.</font color> | |||
===Day 3: Measuring bacterial population growth in lag phase=== | |||
====TO DO:==== | |||
*Innoculate large volumes and collect data for lag phase<br> | |||
[[Image:Note mini.png]]''<font color = red>TEACHERS: ''The procedure assumes each lab group will measure all 4 cultures. Due to equipment constraints, you may wish to vary this experiment so larger "class size" cultures are grown. In that case, increase the solutions and the amount of bacteria added by a factor equal to the number of lab groups. Students can then remove aliquots from these larger cultures for analysis.</font color> | |||
=====Procedure, if using a spectrophotometer===== | |||
1. Prepare a stock growth solution with | |||
*300 ml Luria broth | |||
*300 μl Ampicillin | |||
*250 μl isoamyl alcohol<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''While isoamyl alcohol is safe enough for lab use, it is best if this is added by the teacher.</font><br> | |||
2. Mix this stock growth solution, by swirling the bottle or vortexing gently.<br> | |||
3. Set aside 2 ml of this mixture for each student group into a small sterile culture tube. This aliquot will serve as the blank for the spectrophotometer.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''If you are using a small plastic cuvette, a 1 ml sample will be sufficient.</font color><br> | |||
4. Move 50 ml of the broth solution to 125 ml sterile erlenmeyer flask and add 2ml of bacteria from one of the overnight cultures, e.g. strain 1-1.<br> | |||
5. Repeat the addition of 2ml of bacteria to 50 ml of broth in the erlenmeyer flasks for each of the overnight cultures. <br> | |||
6. Cover the flasks with foil and start them gently stirring on the stir plates.<br> | |||
7. Remove 2 ml from each sample to read the starting density of each. If you are testing all 4 samples you should now have 5 small test tubes (4 with bacterial dilutions and one blank).<br> | |||
8. Prepare the spectrophotometer by setting it to OD600.<br> | |||
9. Note the time and take an "initial" density reading for the bacterial samples. Please note that your teacher may have carried out the preceding steps in advance of the lab. If that is the case, the teacher will tell you how much time has elapsed. That time will be your T<sub>0</sub>.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''Given time constraints in most lab classes, it is OK for the students to prepare these samples and then place them in the refrigerator. The teacher can them place the samples on the room temperature stir plate the next morning and note the time. This may not be necessary during an extended lab period. At room temperature, it will take around three hours for the cultures to enter log phase. Initial readings will have an OD 600 around 0.05.</font><br> | |||
10. Add a stir bar to each culture flask and place onto stir plates. Stir slowly. Cover the flasks with foil.<br> | |||
11. After 20 minutes, remove 1-2 ml from each sample and place in a cuvette. Note: the volume you use here will depend on the size of the cuvette appropriate for your spectrophotometer. Please follow the teacher's instructions. <br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''1 ml is sufficient for a small plastic cuvette, while 2 mls will be needed for a small test tube that fits into most Spec 20 spectrophotometers</font><br> | |||
12. Read the blank and adjust the % Absorbance to zero.<br> | |||
13. Read the sample tubes and record the % Absorbance.<br> | |||
14. Sniff the flask for any evidence of a banana smell, comparing the smell with the banana extract standards. Be sure to shake the standards and the cultures before sniffing. Record your data.<br> | |||
15. At 20 minute intervals repeat steps 11-14.<br> | |||
16. Between time points, you can calculate the bacterial population: 1 OD600 unit = 1 x 10<sup>9</sup> bacteria.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''If the entire growth curve (i.e. days 3, 4 and 5) is to be done in one class, you may have to start the early time points in advance. If you are dividing the growth curve into several short lab periods, be sure to store the cells in the fridge (~4°) until the next session. </font><br> | |||
=====Procedure, if no spectrophotometer is available===== | |||
The turbidity of the bacterial populations can be estimated using the [http://en.wikipedia.org/wiki/McFarland_standards McFarland Turbidity Scale]. This method uses suspensions of a 1% BaCl<sub>2</sub> in 1% H<sub>2</sub>SO<sub>4</sub> that are visually similar to suspensions of various populations of ''E. coli.''<br>[[Image:Turbidity_photo.jpg|thumb|center|400px| Turbidity comparisons for some bacterial cultures (left) and McFarland standards (right)]]<br style="clear:both" /> | |||
1. Following your teacher's instructions, obtain small clear test tubes containing the turbidity standards. The tubes should contain enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom. Make sure each tube is properly labeled with its turbidity standard number. If you are filling the tubes from stock bottles of the standards, use small tubes and place enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom.<br> | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''The size of the tubes and the volume of sample and standard used is flexible. The important things are that the volume can obscure the thick black lines and that the samples and standards are prepared in the same fashion, as shown in the photo. </font color><br> | |||
2. Place the samples in a test tube rack that allows you to view them from the side. Use small tubes and place enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom.<br> | |||
3. On a blank index card or paper use a marker to draw two thick black lines. These lines should be within the height of the standards.<br> | |||
4. Place the card with the lines behind the standards.<br> | |||
5. To compare your bacterial cultures to the standards, you will need to place the bacterial sample in a test tube of the same size and equal volume as the standards. be sure to label these sample tubes.<br> | |||
6. Place the sample tube next to the standard tubes. You should move the sample to compare it to the standard tubes with the most similar turbidity. You can make this assessment more precise by looking for a standard that most similarly obscures the black lines on the background card.<br> | |||
7. Use the table below to determine the comparable OD 600.<br> | |||
8. 1 OD 600 unit equals approximately 1 x 10<sup>9</sup> cells. | |||
[[Image:McFarland_table.PNG]] | |||
===Day 4: Measuring bacterial population growth in log phase=== | |||
====TO DO:==== | |||
=====Restart cultures and collect data for lag phase===== | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''A successful time course for Day 4 could be: Start the samples up in the morning for a class later in the day. These hours will allow the population to reach log phase. At this point, the students can take 2 or 3 more readings, as described for Day 3. The culture can be run overnight at room temperature to reach stationary phase for Day 5</font> | |||
===Day 5: Measuring bacterial population growth in stationary phase=== | |||
====TO DO:==== | |||
=====Collect data for stationary phase===== | |||
[[Image:Note mini.png]]''<font color = red> TEACHERS: ''Two or three readings from each phase will provide sufficient data points for construction of the population growth curve. Just make sure that the students are tracking the time accurately from the initial reading. </font> | |||
==Workflow Protocol B== | |||
===Classroom Content=== | ===Classroom Content=== | ||
[[Image:Intro lab1 BioPrimer1.png|thumb|600 px| [http://openwetware.org/images/3/30/BioPrimerNo1.pdf BioPrimer #1 pdf] ]] | [[Image:Intro lab1 BioPrimer1.png|thumb|600 px| [http://openwetware.org/images/3/30/BioPrimerNo1.pdf BioPrimer #1 pdf] ]] | ||
Revision as of 19:08, 12 July 2011
|
Eau That Smell Lab notes |
PDF of this page
Teacher ConsiderationsThis lab provides a valuable opportunity to teach microbiology techniques, population growth dynamics, molecular genetics and basic synthetic biology concepts in a meaningful, real world way. As can be seen in the discussion questions for the lab report, the analysis of the lab will provide the students with a chance to do meaningful error analysis and examine the difference between quantitative results and qualitative results. This lab offers two different protocols based on the time the teacher wishes to allow. Each of these protocols covers the same concepts but allows for different emphases. Protocol A: This is a shorter procedure for the students. This shorter protocol allows for more emphasis on microbiological techniques. This protocol is recommended should the teacher choose to have the students do the initial bacterial culturing. Essentially, a day prior to any data collection, the large cultures are set up. Instead of letting them all run, part of the culture is immediately removed and placed in the refrigerator. This will be the lag phase sample. After 5-7 hours, a second sample is removed. This will be the log phase sample. The last third of the culture will be allowed to grow overnight. This will be the stationary phase sample. This will allow the students to easily obtain data in one lab period. 1.3 If time allows the students can carry out all parts of the procedure. This will take about 5 class periods in a typical High School AP Biology or Biotechnology class. If instead the teacher prepares in advance the samples as outlined in Part 1 of the procedure, the students can conduct the smell tests and population measurements over two or three days. In a college lab course, most of the bacterial growth curve can be conducted in a typical three to four hour lab period. The procedure includes instructions for using a spectrophotometer to measure the population growth. If a spectrophotometer is not available, the population can be easily measured using the McFarland Turbidity methodology, as explained below. Needed MaterialsTeacher Provides
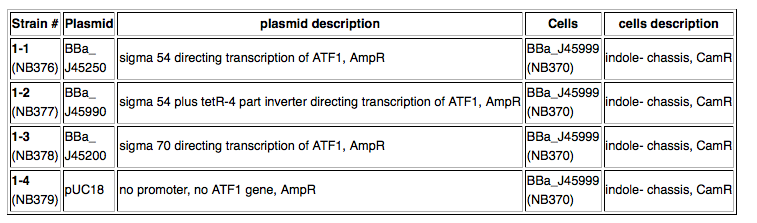
Kit Provides4 strains (see table below)
Chemicals Room Temperature
4° (fridge)
Chemical Hood
Workflow Protocol AClassroom Content
Annotated ProcedureDay 1:TO DO
Streak out strains from stabs to platesWe will be receiving our bacteria with the plasmid already inserted. This culture will come in the form of a "stab" or "slant", a test tube with a small amount of bacteria on a slanted media. To continue the experiment we will have to further culture the bacteria by streaking out the stabs onto LB+amp plates. The plates will be incubated 37° overnight.
This video illustrates the technique used for this transfer. Prepare banana extract standardsThe banana extract is provided in the kit. It will be necessary to make up the standards following the table. Day 2:TO DO:
Grow liquid overnights of bacterial strains1. Using a sterile inoculating loop, transfer a bacterial colony from one of the petri dishes to a large sterile culture tube containing 5 ml of Luria Broth and 5 μl of ampicillin. Prepare turbidity standardsAs the populations of bacteria increase, the culture media gets increasingly turbid. Using the McFarland Turbidity Scale, it is possible to estimate the changes in turbidity. The results will not be as precise as what you would measure with a spectrophotometer, but the changes over time will be detected and the results can be graphed. 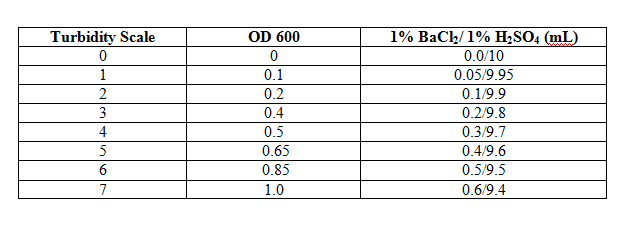
Day 3: Measuring bacterial population growth in lag phaseTO DO:
Procedure, if using a spectrophotometer1. Prepare a stock growth solution with
Procedure, if no spectrophotometer is available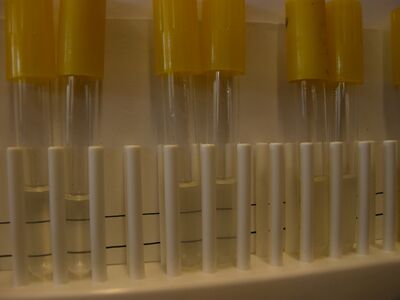 1. Following your teacher's instructions, obtain small clear test tubes containing the turbidity standards. The tubes should contain enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom. Make sure each tube is properly labeled with its turbidity standard number. If you are filling the tubes from stock bottles of the standards, use small tubes and place enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom. Day 4: Measuring bacterial population growth in log phaseTO DO:Restart cultures and collect data for lag phase
Day 5: Measuring bacterial population growth in stationary phaseTO DO:Collect data for stationary phase
Workflow Protocol BClassroom Content
Annotated ProcedureDay 1:TO DO
Streak out strains from stabs to platesWe will be receiving our bacteria with the plasmid already inserted. This culture will come in the form of a "stab" or "slant", a test tube with a small amount of bacteria on a slanted media. To continue the experiment we will have to further culture the bacteria by streaking out the stabs onto LB+amp plates. The plates will be incubated 37° overnight.
This video illustrates the technique used for this transfer. Prepare banana extract standardsThe banana extract is provided in the kit. It will be necessary to make up the standards following the table. Day 2:TO DO:
Grow liquid overnights of bacterial strains1. Using a sterile inoculating loop, transfer a bacterial colony from one of the petri dishes to a large sterile culture tube containing 5 ml of Luria Broth and 5 μl of ampicillin. Prepare turbidity standardsAs the populations of bacteria increase, the culture media gets increasingly turbid. Using the McFarland Turbidity Scale, it is possible to estimate the changes in turbidity. The results will not be as precise as what you would measure with a spectrophotometer, but the changes over time will be detected and the results can be graphed. 
Day 3: Measuring bacterial population growth in lag phaseTO DO:
Procedure, if using a spectrophotometer1. Prepare a stock growth solution with
Procedure, if no spectrophotometer is available 1. Following your teacher's instructions, obtain small clear test tubes containing the turbidity standards. The tubes should contain enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom. Make sure each tube is properly labeled with its turbidity standard number. If you are filling the tubes from stock bottles of the standards, use small tubes and place enough standard in each to fill the tube to a height of about 1 inch (2.5 cm) from the bottom. Day 4: Measuring bacterial population growth in log phaseTO DO:Restart cultures and collect data for lag phase
Day 5: Measuring bacterial population growth in stationary phaseTO DO:Collect data for stationary phase
AssessmentLab Report RubricLab Report ScoreSheetSurvey Monkey LinkTo help us improve the labs, you can send the students here where they can offer anonymous feedback. Thanks! Variations to try
FeedbackWe're always looking to hear back from you if you've thought about this unit, tried it, or stumbled across it and want to know more. Please email us through BioBuilder, info AT biobuilder DOT org. Navigation
|