Uploads by Joseph W. Foley
From OpenWetWare
Jump to navigationJump to search
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | Description |
|---|---|---|---|---|
| 02:47, 15 June 2023 | SPRI buffers v2 3.pdf (file) | 371 KB | typo corrected: 0.5 mL Tween should be 5 mL | |
| 22:13, 11 May 2016 | Library pooling worksheet.ods (file) | 29 KB | made example numbers less ambiguous | |
| 16:51, 9 May 2016 | Illumina qPCR oligos for IDT.txt (file) | 189 bytes | Primer and probe designs for Illumina library qPCR, formatted for IDT purchasing (use the "Bulk Entry" button on the [http://www.idtdna.com/site/order/oligoentry "Custom DNA" interface]). Note that IDT's "/36-TAMSp/" modification includes TAMRA conjugated | |
| 16:49, 9 May 2016 | Illumina qPCR oligos for IDT.tsv (file) | 189 bytes | Primer and probe designs for Illumina library qPCR, formatted for IDT purchasing. Note that IDT's "/36-TAMSp/" modification includes TAMRA conjugated to a dT, so the final T is omitted from the sequence of unmodified bases. | |
| 06:13, 9 May 2016 | Critical step.png (file) | 2 KB | fixed typo | |
| 06:04, 9 May 2016 | Library qPCR example std-crv1.jpg (file) |  |
317 KB | |
| 05:51, 9 May 2016 | Library qPCR example amp-plot1.jpg (file) |  |
370 KB | |
| 22:19, 15 January 2016 | SPRI buffers v2 2.pdf (file) | 486 KB | SPRI buffer protocol (Ludmer Center version 2.2) | |
| 17:52, 16 November 2015 | RNA SPRI validation example.png (file) | 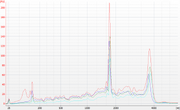 |
99 KB | The original diluted input (red) was loaded with samples purified using different ratios of RNA bead mix: two volumes (blue), one volume (green) and half volume (cyan). The samples were denatured before loading. |
| 17:51, 16 November 2015 | DNA SPRI validation example.png (file) | 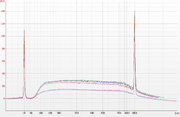 |
433 KB | Validation done with λ phage genomic dsDNA as specified above and loaded on an Agilent Bioanalyzer DNA 1000 chip. Fragmented stock was purified with AMPure XP (3 measurement replicates) and the test sample with homemade bead mix (3 purification replicate |