Pecreaux:Research
 |
|
Goal
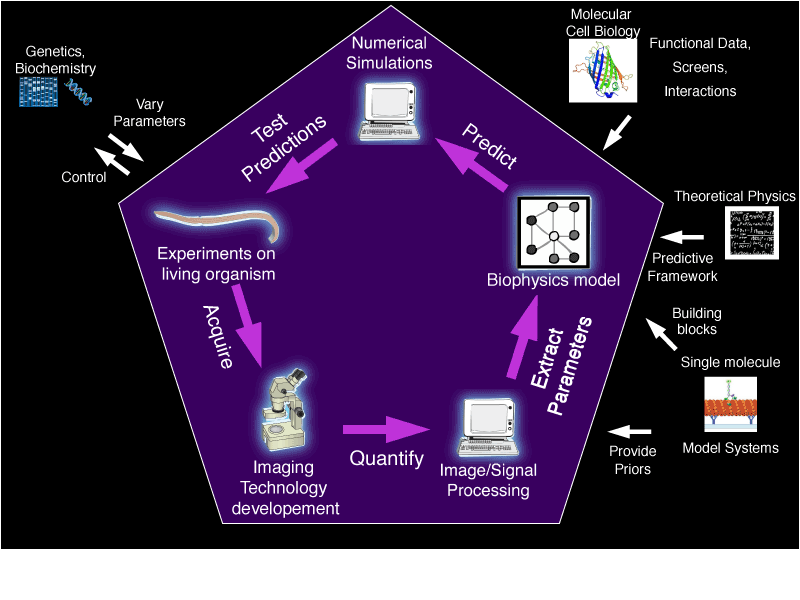
Molecular biology, biochemistry and biophysics in vitro already provides a vast knowledge of individual molecules. Cell biology qualitatively describes mitosis events, while simulations and theoretical approaches propose qualitative understanding at cell scale. However, to quantitatively connect these micro- and macroscopic scales, a bottom-up, engineering approach is yet to be built. Experimental systems biophysics — formulated in the standard language of physics — can offer such a link and account for in vivo interactions, concentrations and biophysics characteristics of the players. We aim to use asymmetric division as a model system to understand the general principles of division because of its rich dynamics and stereotypic cascade of phenomena that facilitates estimation of forces. Starting with the Caenorhabditis elegans embryo, suitable because of its large size, its established cell biology and genetics, we will extend my research to sensory organ precursors in Drosophila melanogaster,
(See also Image processing research at Image Processing)
Approach
To break the current limitations in connecting micro- and macro-scopic scales, we will implement an experimental systems biophysics approach:
- Experimental approaches to center the model on in vivo experimental information (interactions, players’ characteristics, concentrations, etc.) making no speculations or assumptions, and to quantitatively validate the model by comparing model predictions upon parameter variation to protein partial or full depletion phenotypes. Quantifying will require the development of experimental physics and advanced image processing methods.
- Biophysical modeling to extract simple and general rules: “reverse-engineering” the cell mechanism. Designing and analyzing the model will be the opportunity to determine which properties or interactions are essential.
- Systems-level approach will be essential to address the highly complex and regulated nature of cell division, account for robustness to noise, and capture interactions and collective behaviors, by integrating all the players into modeling.
significance and outcome
Experimental systems biophysics, by its ability to reverse engineer mechanisms while keeping connexion with molecular details, and validating models through strict quantitative in vivo assays, can provide novel perspectives in understanding many biological phenomena, efficiently supplementing molecular cell biology and genetics approaches, as well as in vitro or theoretical studies. By its standard language, it will highlight similarities in design, not only across species but also between apparently dissimilar biological phenomena sharing similar molecular players.
Experimental systems biology of cell division will not only be beneficial to fundamental biology, but also to more applied research through its engineering approach, and to all microfilament related fields. From the deciphering of the cell division mechanism, we expect a better understanding of related topics both in basic research like stem cells or applied research. A prominent application is cancer: a deeper understanding of cell division will open new perspectives in understanding how a cell become cancerous in relation to division defects, while the engineering approach will foster the development of targeted cancer therapeutics.
Finally, the algorithms and software developed here will have a broader range of applications: we will make them publicly available to other academic researchers as Fiji/ImageJ plugins. More broadly, the multidisciplinary methods developed for the project can be of inspiration for quantitative biology.