Pecreaux
 |
|
Pecreaux lab's news and job offers may be found on its institutional website: CeDRE team
Latest publication
2023 - Only three principal components account for inter-embryo variability of the spindle length over time. Le Cunff Y., Chesneau L., Pastezeur S., Pinson X., Soler N., Fairbass D., Mercat B., Rodriguez Garci R., Alayan Z., Abdouni A., de Neidhardt G., Costes V., Anjubault M., Bouvrais H., Héligon C. and Pecreaux J. published in BioRxiv.
2022 - Kinetochore microtubules flux poleward along fixed centrosome-anchored microtubules during the metaphase of C. elegans one-cell embryo. Soler N., Chesneau L., Bouvrais H., Pasteuzeur S., Le Marrec L. and Pecreaux J. published in BioRxiv.
2021 - Our research on coordination of spindle-positioning forces during the asymmetric division of the Caenorhabditis elegans zygote published in EMBO reports in may 2021 has been the subject of a vulgarization article on the CNRS website (in french).
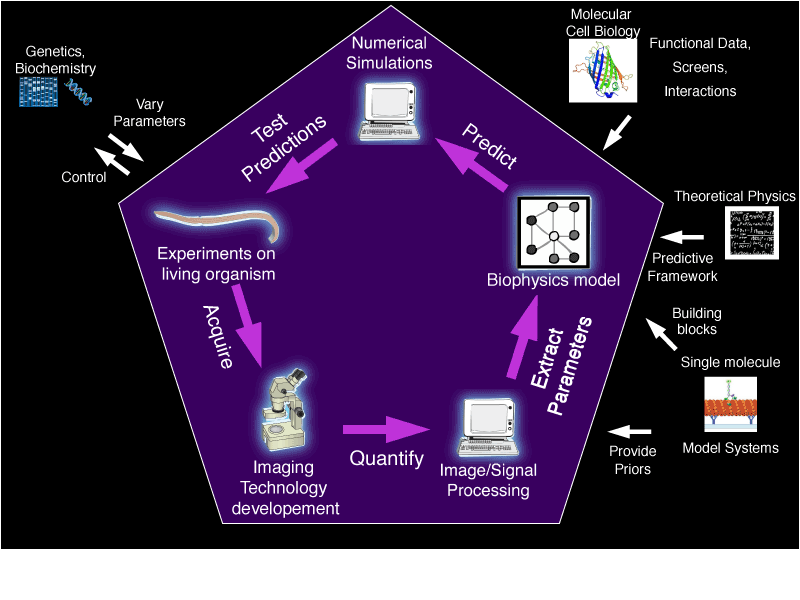
The CeDRE team investigates cell division through a multidisciplinary approach, linking molecular details to cell-level events. Its research interest sums up into the question: How does a robust and adaptive cell division emerge from the numerous interactions of involved players? We hypothesize that the robustness to perturbations (e.g. chromosomal instability in cancer) or adaptability (e.g. to protein evolution) emerges from a network of interacting players well approached by statistical physics. While the proteins differ between organisms, the network is evolutionarily conserved. Such a change of paradigm is highly promising for future applications in cancer therapy.
We focus on in vivo mechanics, i.e. regulation of forces that position the spindle, separate sister chromatids, etc. We investigate not only the dynamics of components but also equilibrium stability. For example, the slow drift in position or length suggest that the spindle is out-of-equilibrium ; it is thought to provide advantage for adaptability. To do so, we develop microscopy and image processing tools to quantify the dynamics in vivo using Caenorhabditis elegans as a model organism and we model the obtained data using out-of-equilibrium statistical physics.
Approach
To achieve such research, our team is multidisciplinary including:
- Biophysical modeling
- featuring a systems approach to understand collective behaviors of the players and robustness to noise; this will enable me to distinguish the dominant mechanisms.
- Validation by in vivo quantitative experiments
- using mainly partial RNA interference, in which the continuous depletion of a protein corresponds to parameter variation in the biophysical model;
- Advanced image processing and experimental physics quantification
- to ensure a detailed comparison to the model’s predictions.
Such an “experimental systems biophysics” approach, by reconciling micro- and macro-scopic scales, can provide novel perspectives in understanding biological phenomena, complementing molecular cell biology approaches, and in vitro or theoretical studies. It is particularly suitable at highlighting collective behaviors involving multiple players (proteins) to create robust cell division, able to compensate for changes in genes or environment for example.