Matthew E. Jurek Week 5
From OpenWetWare
Jump to navigationJump to search
Matthew E. Jurek BIOL398-03/S13
Assignment Page
- BIOL398-03/S13:Week_2
- BIOL398-03/S13:Week 3
- BIOL398-03/S13:Week 4
- BIOL398-03/S13:Week_5
- BIOL398-03/S13:Week_6
- BIOL398-03/S13:Week_8
- BIOL398-03/S13:Week_9
- BIOL398-03/S13:Week_11
- BIOL398-03/S13:Week_12
- BIOL398-03/S13:Week_13
- BIOL398-03/S13:Week_14
User Page
- Matthew E. Jurek Week 2
- Matthew E. Jurek Week 3
- Matthew E. Jurek Week 4
- Matthew E. Jurek Week 5
- Matthew E. Jurek Week 6
- Anthony J. Wavrin/Matthew E. Jurek Metabolic Pathways Project
- Matthew E. Jurek Week 8
- Matthew E. Jurek Week 9
- Matthew E. Jurek Week 11
- Matthew E. Jurek Week 12
- Matthew E. Jurek Week 13
- Matthew E. Jurek Week 14
- Final Presentation
Saccharomyces Genome Database
- Which of these genes has a homolog (similar gene related by descent) in humans? What disease does a deficiency of this gene cause in humans?
- GDH2 has the following homologs in humans: GLUD1 and GLUD2. Deficiencies in these result in hyperinsulinism-hyperammonemia syndrome as well as other neurological disorders. GDH2
- How is the expression of each of these genes regulated?
- GDH1- Expression is regulated by carbon and nitrogen sources. GDH1
- GDH3- Expression is regulated by carbon and nitrogen sources. GDH3
- GDH2- Expression is regulated by nitrogen catabolite repression and intracellular ammonia levels. GDH2
- GLN1- Expression is regulated by nitrogen source and by amino acid limitation. GLN1
- GLT1- Expression is regulated by nitrogen source. GLT1
- Using the compound search tool of SGD, search on "L-glutamate". How many pathways does it participate in?
- "L-glutamate" participates in 26 pathways as a reactant and 30 as a product. It is also found in a number of reactions that have not yet been assigned a pathway. L-glutamate
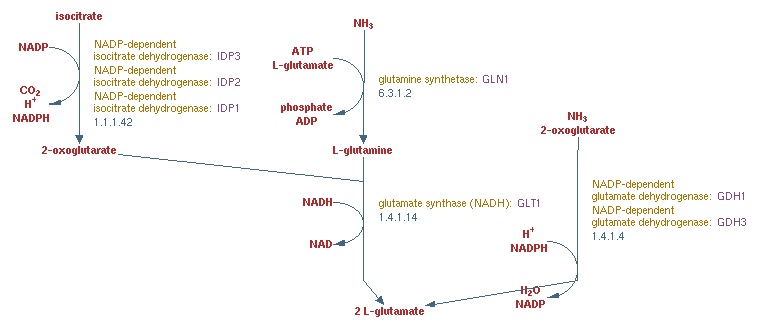
- Find the SGD representation of the pathway we are working on in class and attach a screenshot and hyperlink to your journal page. Choose the one that shows all of the reactions we talked about in class and make sure you can relate it to your notes, matching the genes, enzyme names, and reactants/products.
via Superpathway of glutamate biosynthesis
- What parameters for these reactions can you find using this database?
- There are numerous reference articles for each gene listing numerous parameters. There is a definite overlap regarding the parameters in the references and the parameters in the ter Schure paper. This includes things like carbon, nitrogen, ammonia and biomass.
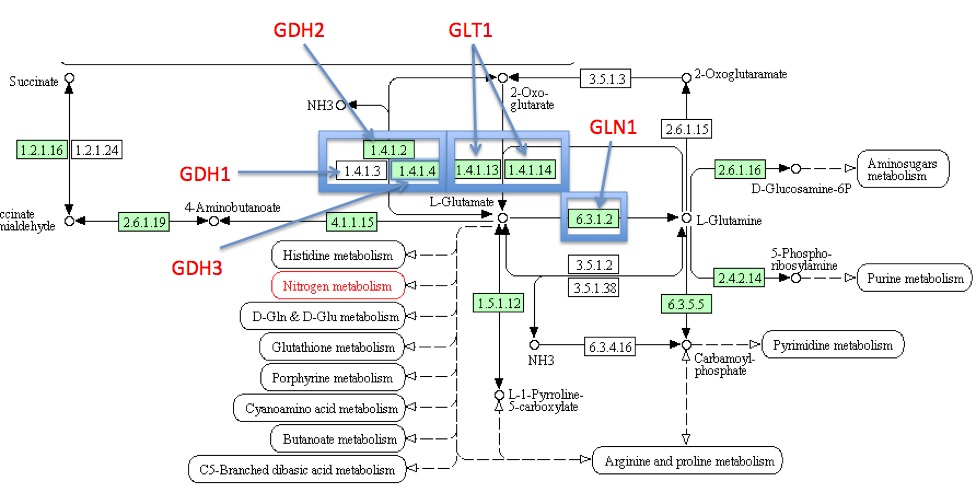
KEGG
via KEGG Database]
- How many genes in this pathway exist in yeast?
- There are 12 highlighted genes, meaning these 12 exist in yeast.
- Click on each of the five enzymes of our pathway and read the individual enzyme pages. Is there any new information here that was not represented by SGD?
- KEGG includes the amino acid and nucleotide sequence in addition to the information found on SGD.
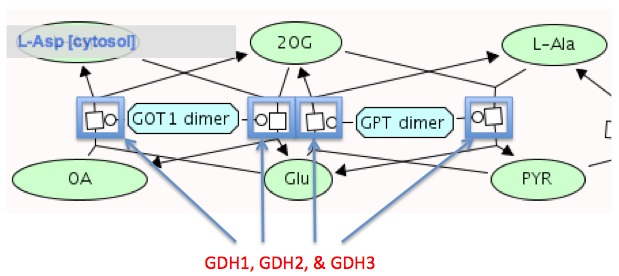
Reactome
- After exploring this database for awhile, it was hard to find our exact pathway. The following image is the closest I could find to what we've been studying. Moving the cursor over the small square and circle between each reaction resulted in a small box. The small box showed the reversible reaction of alpha-ketoglutarate going to glutamate and vice versa. Thus, GDH1, GDH2, and GDH3 are all labeled. Each box showed this reaction in either the forward process or the reverse explaining why the same genes are labeled for every box.
via: Reactome