Mass Univariate ERP Toolbox: creating GND variables
Home/Download Tutorial Revision History Contact/Questions Appendix Other EEG Analysis Software Packages
Tutorial Sections
- Installing the Mass Univariate Toolbox
- Creating GND variables from EEGLAB set or ERPLAB erp files<-You are here
- Within-subject t-tests
- Between-subject t-tests
- General advice
Creating GND variables from EEGLAB set or ERPLAB erp files
The first step towards using the Mass Univariate Toolbox to perform mass univariate analyses is to compute ERPs from each individual subject's data and to store them in a MATLAB struct variable called GND. There are currently two ways to create a GND variable:
- From EEGLAB set files using the function sets2GND.m. Note, currently the Mass Univariate Toolbox expects all EEGLAB set files to contain epoched data. To learn how to convert continuous data into epoched data see the EEGLAB tutorial.
- From ERPLAB erp files using the function erplab2GND.m
1: Creating a GND variable from EEGLAB set files (without ERPLAB)
The Mass Univariate Toolbox uses the concept of “bins” to create ERPs. A bin is simply a class of event to which one would like compute an ERP. For example, consider a simple oddball experiment in which participants see two types of stimuli, Targets and Standards. Bin 1 might consist of all the target stimuli and Bin 2 might consist of all the standards. To format one’s data for use by the Mass Univariate Toolbox, one needs to:
- Create a “bin list file” which specifies which event types in your set file belong in which bins
- Use the toolbox function bin_info2EEG.m to add bin events to EEGLAB set files. Specifically, they are added to the fields EEG.event and EEG.epoch of the EEG variable
The following example shows how to add bins to data from four participants in a simple N400 experiment. In this experiment, participants listened to words preceded by either a synonym or a non-synonym. You can download the data set here: demo N400 set files
The data were taken from the EEGLAB wiki and have been modified so that now some independent components in the files are marked as EEG artifacts (e.g., blinks, muscle activity).
Note, when adding bin information to EEGLAB set files and creating GND variables, it's probably a good idea to keep the EEGLAB GUI closed to prevent interference between the two.
1.1: Create a bin list file
A bin list file is simply a text file in which each line specifies a type of event that belongs to a particular bin. More specifically, each line is composed of the following three elements:
- bin_number
- event_type
- bin_descriptor
arranged in order. For example, the following line means that events of type S253 fall in Bin #1, which consists of Synonyms:
1) S253=Synonyms
When more than one type of event falls in the same bin, you can use multiple lines to capture all the event types. For example, the following lines would mean that events of type S253 and S200 fall in Bin #1, which consists of Synonyms:
1) S253=Synonyms
1) S200=Synonyms
Or you could combine those two lines into a single line:
1) S200 S253=Synonyms
If the events are numeric and consecutive you can specify multiple types of events per line using the MATLAB colon convention. For example, the following lines would mean that events of type 200, "201", and "202" fall in Bin #1, which consists of Synonyms:
1) 200:202=Synonyms
The bin list file for the example N400 data set can be downloaded here: bin list file example. Note that:
- The bin numbers need to be integers running from 1 to n (where n is the total number of bins).
- The event types (e.g., "S253") must already be present in the set file, specifically, as character strings in the EEG.epoch(#).eventtype = {...} cell array. You can use the function list_event_types.m to tell you which event types are in a particular set file. For example, to list the event types in the first example N400 data set, enter:
>>[types type_count]=list_event_types('syn02-s253-clean.set');
1.2: Add bin event types to EEGLAB set files
Once you've created a bin list file, you can use it to add bin events to a set file via the toolbox function bin_info2EEG.m.
Note, if you have multiple set files per subject, you need to put the subject's codename in the EEG.subject field of the EEG variable. If EEG.subject is empty, the toolbox will assume that the set filename is the subject's codename.
Currently, bin_info2EEG.m only allows you to add bin information one set file at a time. For example, to add bin types to the set file syn02-s253-clean.set (in the directory s02 in the demo N400 data set) do the following:
>> bin_info2EEG('syn02-s253-clean.set','synVSnonsyn_blf.txt','syn02-s253-clean.set');
You should get the following output at the MATLAB command line:
'pop_loadset(): loading file syn02-s253-clean.set ...
Reading float file 'syn02-s253-clean.dat'...
'Scaling components to RMS microvolt
Bin 1: Synonyms
Event types belonging to this bin: 'S253'
Bin 2: Nonsynonyms
Event types belonging to this bin: 'S254'
Number of events per bin.
Bin 1 (Synonyms): 231
Bin 2 (Nonsynonyms): 0
Saving EEG variable as the file: syn02-s253-clean.set
Scaling components to RMS microvolt
Saving dataset...
Note that this set file only contains epochs time locked to synonyms.
The bin_info2EEG function scans each epoch in the specified EEG data structure looking for strings in the EEG.epoch(#).eventtype field that match the event type specified in the bin list text file. When matches are found, the function updates EEG.event and EEG.epoch data structures to add a 'bin#' event type at the same latency as the matching event. In addition, the bin descriptor strings for matching event types are assembled into a cell array (in the order given by the bin numbers) and this cell array is added as a new field to the EEG data structure, EEG.bindesc.
Since bin_info2EEG can only handle one set file at a time, it can help to write loops to add bin information to multiple set files at once. For example, the following two loops will finish adding bin information to the remainder of the demo N400 set files:
>> for sub=[5 7 8 10],
fname=['syn' num2str(sub,'%.2d') '-s253-clean.set'];
bin_info2EEG(fname,'synVSnonsyn_blf.txt',fname);
end
>> for sub=[2 5 7 8 10],
fname=['syn' num2str(sub,'%.2d') '-s254-clean.set'];
bin_info2EEG(fname,'synVSnonsyn_blf.txt',fname);
end
1.3: Create a GND variable from your EEGLAB set files
Once bin types have been added to all of your set files, the next step is to compute ERPs for each subject and each bin and to store them in a variable called GND (for "grand average"). You do this with the function sets2GND.m. For example, to make a GND variable from the demo N400 data, you could enter the following command into the MATLAB command line:
>> GND=sets2GND('gui','bsln',[-100 0],'exclude_chans',{'LO1','IO1','SO1','LO2'});
Note that we are choosing not to import four ocular channels (LO1, IO1, SO1, and LO2) because we don't have coordinates for them.
A GUI will then open to ask you what set file files you would like to include in your GND variable:

After you've selected some files, you will be asked if you want to select any more files (e.g., files in a different directory):
Once you're done selecting files, MATLAB will begin collecting trials for each subject. For each set file, something like the following should appear at the MATLAB command line:
pop_loadset(): loading file /Users/dgroppe/Documents/MATLAB/DEMO_DATA/5subjects_post_groppe/sub02_nonsyn.set ...
Scaling components to RMS microvolt
Scaling components to RMS microvolt
Removing artifact ICs from EEG set file: S2-1-2 S254-clean
19 total artifact ICs will be removed.
Baselining data by removing mean EEG between -100 and 0 ms (time points 401 and 421).
Finally, once all the set files have been loaded, a summary of the number of trials found for each bin and subject is displayed on the MATLAB command line:
Trials per bin for Subject S02:
Bin 1 (Synonyms): 231 trials
Bin 2 (Nonsynonyms): 223 trials
Trials per bin for Subject S05:
Bin 1 (Synonyms): 232 trials
Bin 2 (Nonsynonyms): 235 trials
Trials per bin for Subject S07:
Bin 1 (Synonyms): 226 trials
Bin 2 (Nonsynonyms): 220 trials
Trials per bin for Subject S08:
Bin 1 (Synonyms): 227 trials
Bin 2 (Nonsynonyms): 225 trials
Trials per bin for Subject S10:
Bin 1 (Synonyms): 196 trials
Bin 2 (Nonsynonyms): 196 trials
Finally, a GUI will open to ask you what filename you would like to use to store your GND variable:

By convention, GND variables are saved with the extension ".GND" even though they are really MATLAB ".mat" files (just like EEGLAB ".set" files are really MATLAB ".mat" files). So if you ever want to load a GND variable into MATLAB you simply need to type something like the following:
>> load synVSnonsyn.GND -MAT
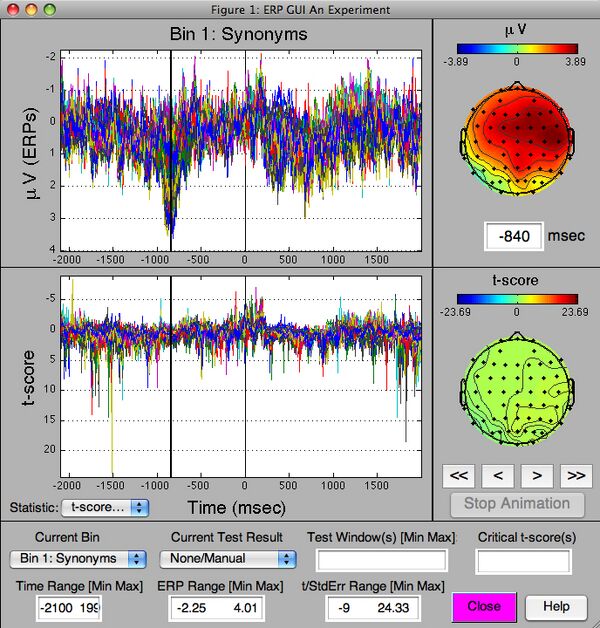
Now that you have your GND variable, you can visualize the ERPs in it via the the ERP GUI like so:
>> gui_erp(GND)
The following GUI should be created. If you hold the mouse over a button or field, a help message should appear that explains what it does.
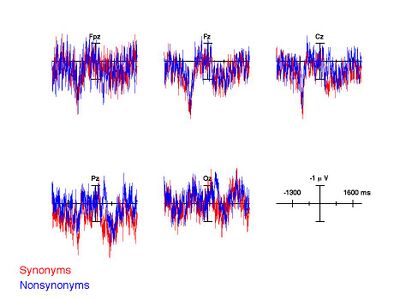
Alternatively, you can visualize the ERPs from multiple bins simultaneously via the function plot_wave.m. We'll just plot a few channels to avoid visual clutter:
>> plot_wave(GND,[1 2],'include_chans',{'FPz','Fz','Cz','Pz','Oz'});
Note, sets2GND.m has a few options not covered here that might be of use. Enter the following to see full documentation on how to use the function:
>>help sets2GND
2: Creating a GND variable from ERPLAB erp files
As mentioned above, the Mass Univariate Toolbox uses the concept of “bins” to create ERPs. ERPLAB also uses the concept of bins to create ERPs and stores ERPs in erp files (see the section of the ERPLAB tutorial on computing ERPs). Thus it is very straightforward to construct a GND variable from a collection of ERPLAB erp files by using the Mass Univariate Toolbox function erplab2GND.m. To illustrate, we show below how to create a GND variable from two erp files, which you can download here. The first erp file was derived from the ERPLAB demo data set and the second erp file is simply a noisy copy of the first for purposes of illustration only.
Once you've downloaded and unzipped the demo erp files, enter the following command into the MATLAB command line:
>> GND=erplab2GND('gui','exclude_chans',{'VEOG','HEOG','A2'});
The input 'gui' specifies that we want to use a GUI to select which erp files to import while the optional input 'exclude_chans' indicates that we do not want to import three channels of data ("VEOG", "HEOG", "A2"). Those three channels are used primarily for artifact detection or re-referencing data and are not of much interest to analyze.
After you enter that command, a GUI like the following should be created for selecting erp files. Select the two demo erp files.
After you've selected some files, you will be asked if you want to select any more files (e.g., files in a different directory). If you've already selected the two demo erp files, click "No."

The following output should appear in the MATLAB command window:
ERP file, /Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s1_with_locs.erp, does not have a subject name. I will use the filename as the subject's name.
Baselining data by removing mean EEG between -100 and -2 ms (time points 1 and 50).
ERP file, /Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s2_with_locs.erp, does not have a subject name. I will use the filename as the subject's name.
Baselining data by removing mean EEG between -100 and -2 ms (time points 1 and 50).
Trials per bin for Subject /Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s1_with_locs.erp:
Bin 1 (Frequent Category (Digit)): 175 trials
Bin 2 (Rare Category (Letter)): 48 trials
Trials per bin for Subject /Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s2_with_locs.erp:
Bin 1 (Frequent Category (Digit)): 175 trials
Bin 2 (Rare Category (Letter)): 48 trials
Baselining data from -100 to -2 ms (that's time point 1 to 50).
Baselining data from Participant #1 (/Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s1_with_locs.erp).
Baselining data from Participant #2 (/Users/dgroppe/Documents/MATLAB/ERPLAB_TUT_DATA/s2_with_locs.erp).
Then you will prompted to name the file that the new GND variable will be stored as:
By convention, GND variables are saved with the extension ".GND" even though they are really MATLAB ".mat" files (just like ERPLAB ".erp" files are really MATLAB ".mat" files). So if you ever want to load a GND variable into MATLAB you simply need to type something like the following:
>> load dem_odbl.GND -MAT
Now that you have your GND variable, you can visualize the ERPs in it via the the ERP GUI like so:
>> gui_erp(GND)
The following GUI should be created. If you hold the mouse over a button or field, a help message should appear that explains what it does.

Note, erplab2GND.m has a few options not covered here that might be of use. Enter the following to see full documentation on how to use the function:
>>help erplab2GND
3: Fields of your GND variable explained
Generally speaking, GND variables contain grand average ERPs, individual participant ERPs, the results of t-tests, and associated information (e.g., electrode locations). Note, some fields of the GND variable (e.g., GND.cals) are only relevant to data from Kutaslab and can be ignored when using the toolbox on data collected from other labs. You can see exactly what's stored in the variable by typing "GND" into the MATLAB command line:
>> GND
GND =
exp_desc: 'An Experiment'
filename: 'synVSnonsyn.GND'
filepath: '/Users/dgroppe/Documents/MATLAB/DEMO_DATA/5subjects_post_groppe/'
saved: 'yes'
grands: [57x820x2 double]
grands_stder: [57x820x2 double]
grands_t: [57x820x2 double]
sub_ct: [4 4]
chanlocs: [1x57 struct]
bin_info: [1x2 struct]
condesc: {'Experiment (not cal pulses)'}
time_pts: [1x820 double]
bsln_wind: [-100 0]
odelay: []
srate: 200
indiv_fnames: {1x8 cell}
indiv_subnames: {'S02' 'S05' 'S07' 'S10'}
indiv_traits: []
indiv_bin_ct: [4x2 double]
indiv_bin_raw_ct: [4x2 double]
indiv_erps: [4-D double]
indiv_art_ics: {1x8 cell}
cals: []
history: []
t_tests: []
Here's what the various fields store:
- exp_desc:<-the name of the experiment
- filename:<-the filename of the GND variable
- filepath:<-the filepath of the GND variable
- saved: <-whether or not the GND variable has been saved since it was last modified
- grands: <-grand average ERPs (channel x time point x bin) in microvolts
- grands_stder: <-standard error of the grand average ERPs (channel x time point x bin) in microvolts
- grands_t: <-t-scores of the grand average ERPs (channel x time point x bin), which is simply the grand average divided by the standard error
- sub_ct: <-the number of subjects contributing to the grand average ERP for each bin
- chanlocs: <-electrode coordinates in EEGLAB format
- bin_info: <-struct array containing the text definition of each bin and it's "condition code." Condition codes are specific to Kutaslab data and can be ignored by non-Kutaslab users.
- condesc: <-text descriptions of each condition code. Again, condition codes are specific to Kutaslab data and can be ignored by non-Kutaslab users.
- time_pts: <-the peri-event time that each ERP time point corresponds to (in milliseconds)
- bsln_wind: <-the time window used to baseline the ERPs (in milliseconds)
- odelay: <-A field specific to Kutaslab data that can be ignored by non-Kutaslab users.
- srate: <-Data sampling rate (in Hz)
- indiv_fnames:<-The filenames of the set files used to produce the GND variable.
- indiv_subnames:<-The codenames of each subject
- indiv_traits: <-A field where you could store traits of individual subjects (e.g., age, working memory span)
- indiv_bin_ct: <-The number of trials each participant contributed to each bin.
- indiv_bin_raw_ct: <-A field specific to Kutaslab data that can be ignored by non-Kutaslab users.
- indiv_erps: <-The ERPs from each individual subject (channel x time point x bin x subject) in microvolts
- indiv_art_ics: <-The numeric indices of independent components removed from each set file before using them to compute ERPs
- cals: <-A field specific to Kutaslab data that can be ignored by non-Kutaslab users.
- history: <-A record of commands applied to the GND variable
- t_tests: <-The results of mass univariate t-test analyses applied to this GND variable
A more readable way to know the basic contents of a GND variable is to use the function headinfo.m ("headinfo" is short for header information):
>> headinfo(GND)
CHANNELS
1:Fp1 2:Fpz 3:Fp2 4:AF3 5:AF4 6:F7 7:F5 8:F3
9:F1 10:Fz 11:F2 12:F4 13:F6 14:F8 15:FC5 16:FC3
17:FC1 18:FCz 19:FC2 20:FC4 21:FC6 22:T7 23:C5 24:C3
25:C1 26:Cz 27:C2 28:C4 29:C6 30:T8 31:CP1 32:CPz
33:CP2 34:CP4 35:CP6 36:TP8 37:P7 38:P5 39:P3 40:P1
41:Pz 42:P2 43:P4 44:P6 45:P8 46:PO5 47:PO3 48:PO1
49:POz 50:PO2 51:PO4 52:PO6 53:O1 54:Oz 55:O2 56:TP7
57:CP5
BINS
Bin 1: Synonyms
Bin 2: Nonsynonyms
t-TESTS
No t-test results have been stored with these data.
CONTINUE ON TO THE NEXT SECTION OF THE TUTORIAL
