Jimenez-Gomez Lab:Research
The Jiménez-Gómez group focuses on the application of modern genetic and bioinformatic techniques to study plant natural variation, evolution and domestication.
There is a vast diversity of environmental conditions on earth, and plants have developed strategies to survive in most of these settings. In addition, artificial selection has shaped the behaviour of particular species to serve human interests. In our lab, we try to identify the molecular mechanisms that allow plants to successfully adapt to this diversity of situations.

There are more than 300,000 plant species living in most places and seasons on earth, which display a plethora of strategies to grow, survive and reproduce. One of the most exciting fields in plant molecular biology uses molecular, genomic and genetic tools to identify the genetic basis of natural variation. However, in their majority these tools have been developed for a few model systems species such as the thale cress, Arabidopsis thaliana. Studies in this Brassicaceae led to the current understanding of molecular biology in plants and allowed the discovery of genes and pathways important for adaptation. But the studies performed with this species have also raised new questions: Do all plants function similarly at the molecular level? What are the genes that allow other species to survive in environments where Arabidopsis is not found? Are the genes important for adaptation in Arabidopsis also important for adaptation in other species?
Tomato is a great system for evolutionary studies due to its history, and has very different characteristics from Arabidopsis. Although its place of origin is in the west part of South America, the first evidences of domestication are found in Mexico. European conquerors brought this plant to Spain, from where it spread to the rest of Europe. Since then tomato has undergone strong artificial selection to become one of the most popular crops in the world. Interestingly, in South and Central America a variety of wild relatives of tomato can be still be found in many diverse habitats such as islands, deserts, high mountains or humid forests (Fig 1). Our interest is to use these species to identify the genes involved in adaptation to all these environments, including the genes that have been selected by humans to transform the wild tomatoes to the crop that we know today.
In non-model species like tomato, exploring the genetic basis of the phenotypic diversity was restricted until recently to the analysis of physiological characteristics and a few localized genomic sequences, genes and proteins. During the last few decades, scientists have used these tools combined with intelligent experimental designs and statistical methods to localize the chromosomal regions that control diversity using association studies and QTL analyses (for example Jimenez-Gomez et al, 2007). Unfortunately, the identification of the genes responsible for this diversity is a difficult task, and the most common result is to end up with hundreds or thousands of candidate genes.
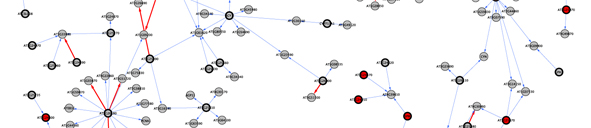
Our lab is interested in implementing bioinformatic tools to help identify the genes underlying these association studies (Jimenez-Gomez & Maloof, 2009). Previous works showed that identification of candidate genes for QTLs could benefit from methods that integrate information from different sources (Jimenez-Gomez et al, 2010)(Fig 2). In addition, it is becoming clear that the reductionist approaches used until now in molecular biology do not represent the real complex architectures found in living cells. Therefore, system biology analyses have the potential to uncover the genes and pathways controlling phenotypes of interest.
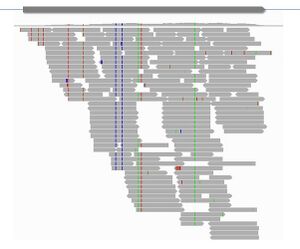
Recent advances in high throughput sequencing have facilitated molecular studies in non-model species. The use of this technology has allowed a consortium of researchers to release the genome of cultivated tomato, and very soon also from a couple of its wild relatives. As in Arabidopsis, genomic sequences will open the door to natural variation studies in tomato. The powerful sequencing technologies also allow acquisition of molecular traits that were only accessible in model organisms, such as gene expression profiling (Fig 3). Moreover, with this technology we can investigate phenomena that are still not well understood even in model systems, like epigenetic modifications, small RNA regulation, copy number variation, allele specific expression, alternative splicing or antisense transcription. We count in the lab with pipelines that allow us to reliably obtain polymorphism and expression information from RNA-seq data from different species. We are interested in developing similar methods to quantify and classify the abovementioned molecular events. The final goal is to associate these molecular traits to the phenotypic variation found in plants.
