Janelle N. Ruiz Assignment 8
Working with Protein Sequences In-class Activity
Task 1
Chapter 2: Retrieving Protein Sequences/Retrieving a list of Related protein sequences (pp. 42-51 in second edition). The example worked through in the book uses the sequence of an enzyme called dUTPase. Follow the book example yourself and then work through the example again, this time using the HIV gp120 envelope protein instead
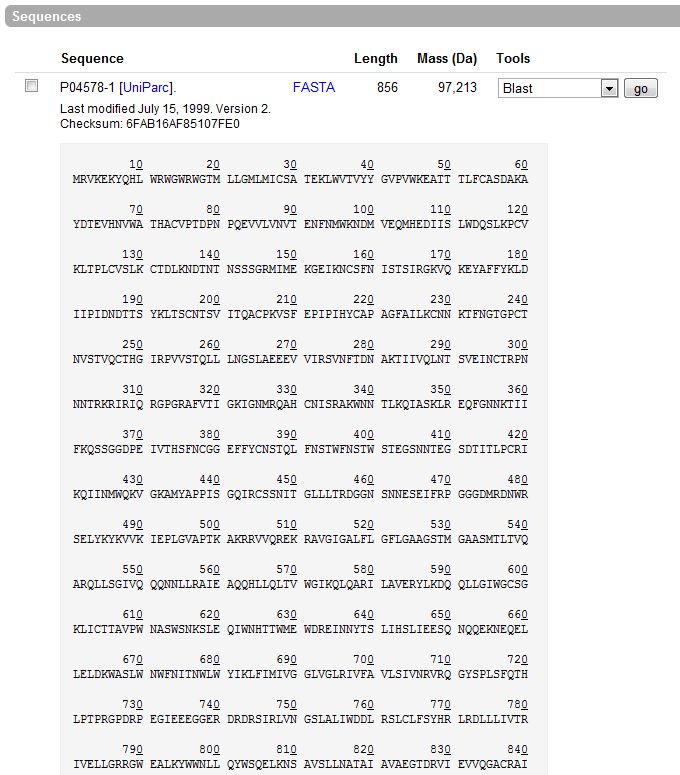
- We typed in “HIV gp120 envelope protein”, eliminating TrEMBL unsupervised computer translations, and obtained four pages of results. We selected P04578, ENV_HV1H2, Human immunodeficiency virus type 1 (isolate HXB2 group M subtype B) (HIV-1), to view amino acid sequence using FASTA formatting.
- >sp|P04578|ENV_HV1H2 Envelope glycoprotein gp160 OS=Human immunodeficiency virus type 1 (isolate HXB2 group M subtype B) GN=env PE=1 SV=2:
- <MRVKEKYQHLWRWGWRWGTMLLGMLMICSATEKLWVTVYYGVPVWKEATTTLFCASDAKAYDTEVHNVWATHACVPTDPNPQEVVLVNVTENFNMWKNDMVEQMHEDIISLWDQSLKPCVKLTPLCVSLKCTDLKNDTNTNSSSGRMIMEKGEIKNCSFNISTSIRGKVQKEYAFFYKLDIIPIDNDTTSYKLTSCNTSVITQACPKVSFEPIPIHYCAPAGFAILKCNNKTFNGTGPCTNVSTVQCTHGIRPVVSTQLLLNGSLAEEEVVIRSVNFTDNAKTIIVQLNTSVEINCTRPNNNTRKRIRIQRGPGRAFVTIGKIGNMRQAHCNISRAKWNNTLKQIASKLREQFGNNKTIIFKQSSGGDPEIVTHSFNCGGEFFYCNSTQLFNSTWFNSTWSTEGSNNTEGSDTITLPCRIKQIINMWQKVGKAMYAPPISGQIRCSSNITGLLLTRDGGNSNNESEIFRPGGGDMRDNWRSELYKYKVVKIEPLGVAPTKAKRRVVQREKRAVGIGALFLGFLGAAGSTMGAASMTLTVQARQLLSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKDQQLLGIWGCSGKLICTTAVPWNASWSNKSLEQIWNHTTWMEWDREINNYTSLIHSLIEESQNQQEKNEQELLELDKWASLWNWFNITNWLWYIKLFIMIVGGLVGLRIVFAVLSIVNRVRQGYSPLSFQTHLPTPRGPDRPEGIEEEGGERDRDRSIRLVNGSLALIWDDLRSLCLFSYHRLRDLLLIVTRIVELLGRRGWEALKYWWNLLQYWSQELKNSAVSLLNATAIAVAEGTDRVIEVVQGACRAIRHIPRRIRQGLERILL>
Task 2
Chapter 4: Reading a SWISS-PROT entry (pp. 110-123 in the second edition). The example worked through in the book is the epidermal growth factor receptor. Work through this example and then do it again with the HIV gp120 envelope protein instead
General Information about the Entry
- Entry Name: ENV_HV1H2
- Primary Accession Number: P04578
- Secondary Accession Numbers: O09779
- Integrated into Swiss-Prot on: August 13, 1987
- Sequence was last updated on: July 15, 1999
- Annotations were last modified on: March 2, 2010
Name and Origin of the Protein:
- Protein Name: Envelope glycoprotein gp160
- Alternative Names: Env polyprotein
- Gene Name: env
- Virus Host: Homo sapiens (Human) [TaxID: 9606]
- Taxonomy: Viruses › Retro-transcribing viruses › Retroviridae › Orthoretrovirinae › Lentivirus › Primate lentivirus group
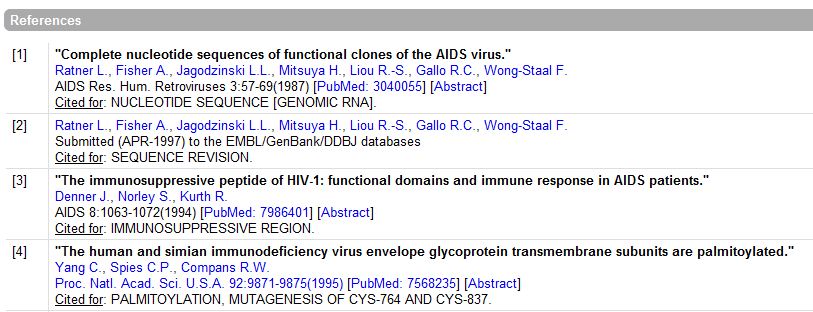
References:
- A sample of references shown:
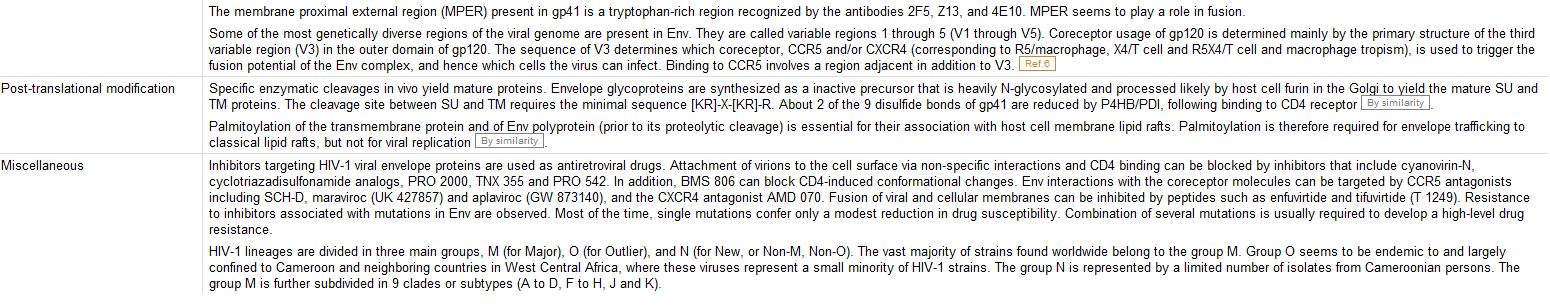
Comments:
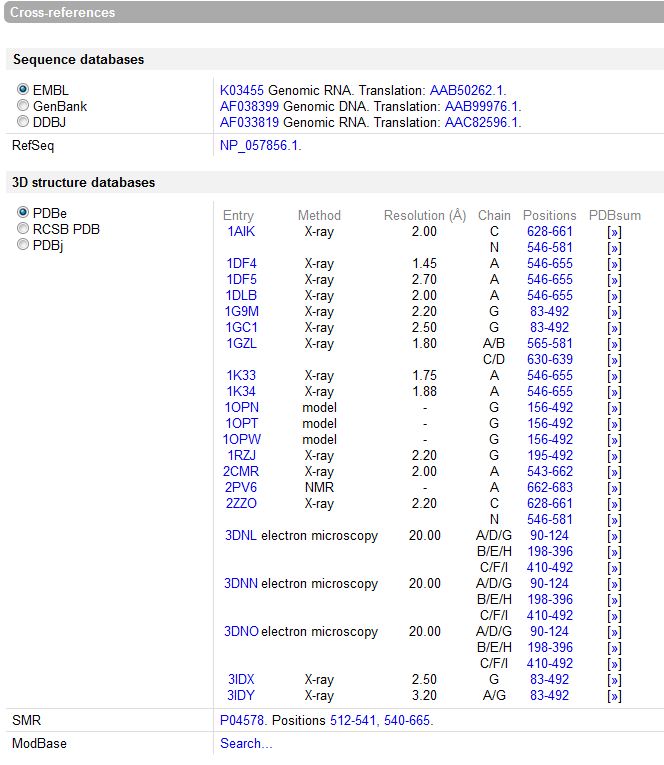
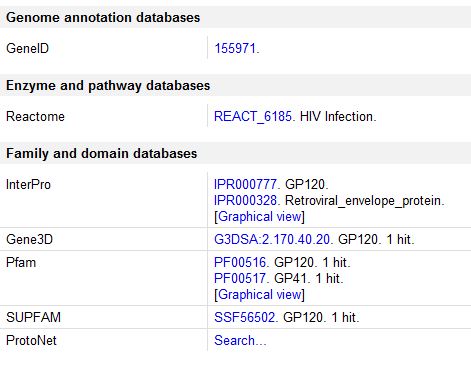
Cross-References:
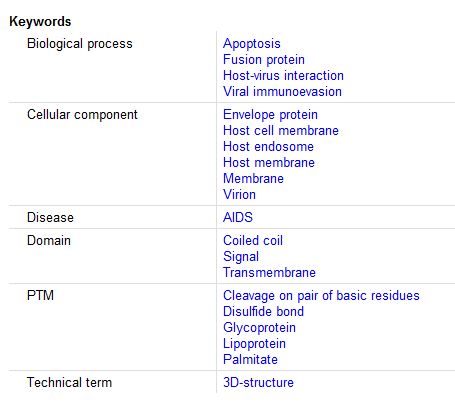
Keywords:
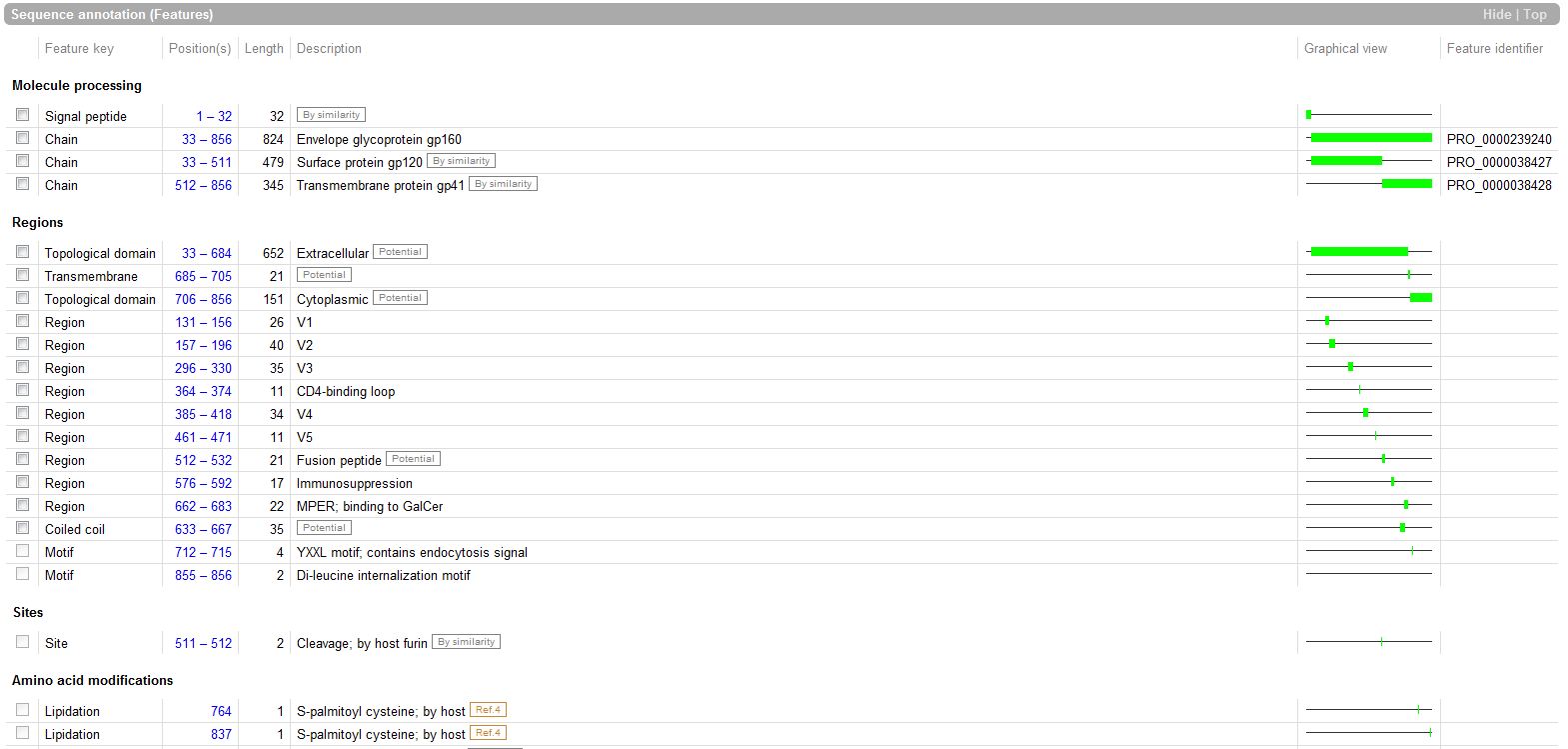
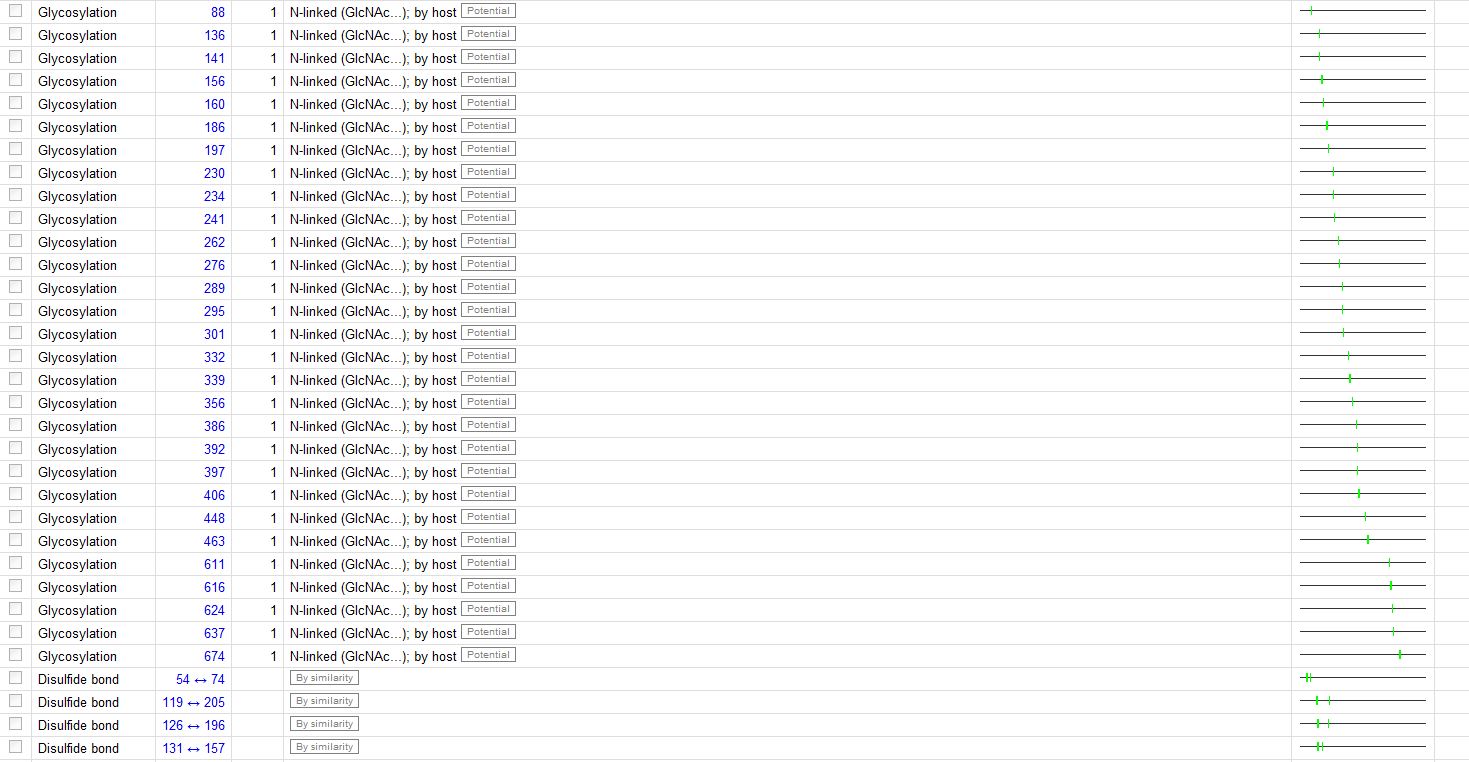
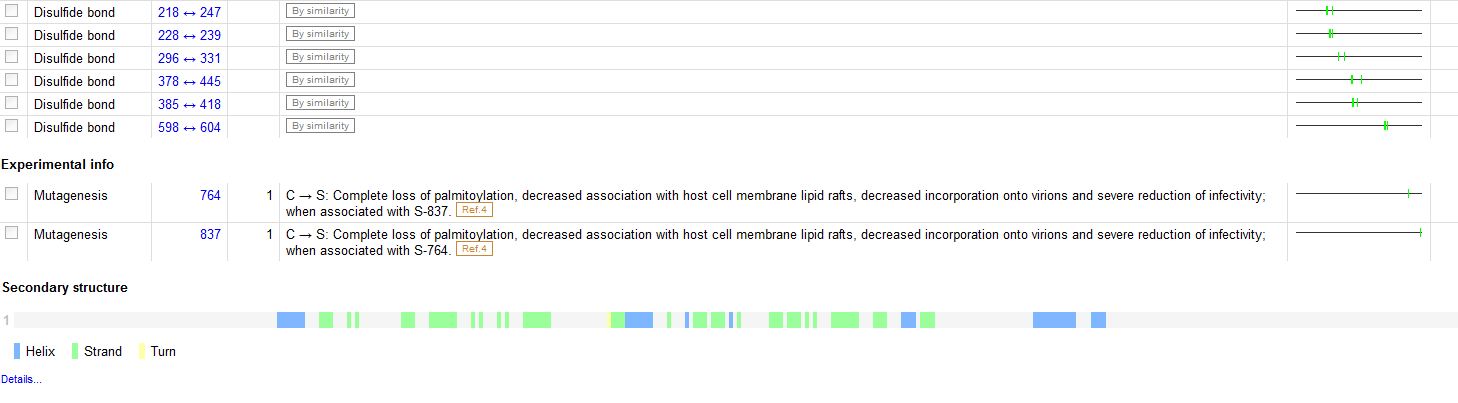
The Features:
The Sequence:
Task 3
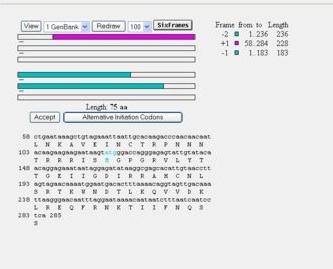
Chapter 5: ORFing your DNA sequence (pp. 146-147 in second edition). In the previous section of the course, we were working with DNA sequences from the HIV gp120 envelope protein. Take one of your DNA sequences and follow the instructions to find the open reading frames in the sequence. Since you were working with just a portion of the entire envelope protein, you may get some strange results. Compare your results with the SWISS-PROT entry you found for the protein above to decipher what the output means. Besides the NCBI Open Reading Frame Finder described in the book, ExPASy also has a translation tool you can use, found here
- Subject 4, "Linear progressor"
- The ORF sequence was similar to SWISS-PROT entry, with some slight differences in sequence.
Task 4
Chapter 6: Working with a single protein sequence (pp. 159-195 in second edition). Work through the following examples in this chapter using the entire HIV gp120 envelope protein sequence that you obtained from SWISS-PROT. We will then compare the results of these analyses with the actual structure of the gp120 protein obtained by X-ray crystallography
ProtParam
Analyzed entire sequence
- Molecular Weight: 97212.7
- Extinction Coefficients:
Extinction coefficients are in units of M-1 cm-1, at 280 nm measured in water.
Ext. coefficient 200185 Abs 0.1% (=1 g/l) 2.059, assuming all pairs of Cys residues form cystines
Ext. coefficient 198810 Abs 0.1% (=1 g/l) 2.045, assuming all Cys residues are reduced
- Instability:
The instability index (II) is computed to be 39.92 This classifies the protein as stable.
- Half-Life:
The N-terminal of the sequence considered is M (Met).
The estimated half-life is: 30 hours (mammalian reticulocytes, in vitro). 20 hours (yeast, in vivo). 10 hours (Escherichia coli, in vivo).
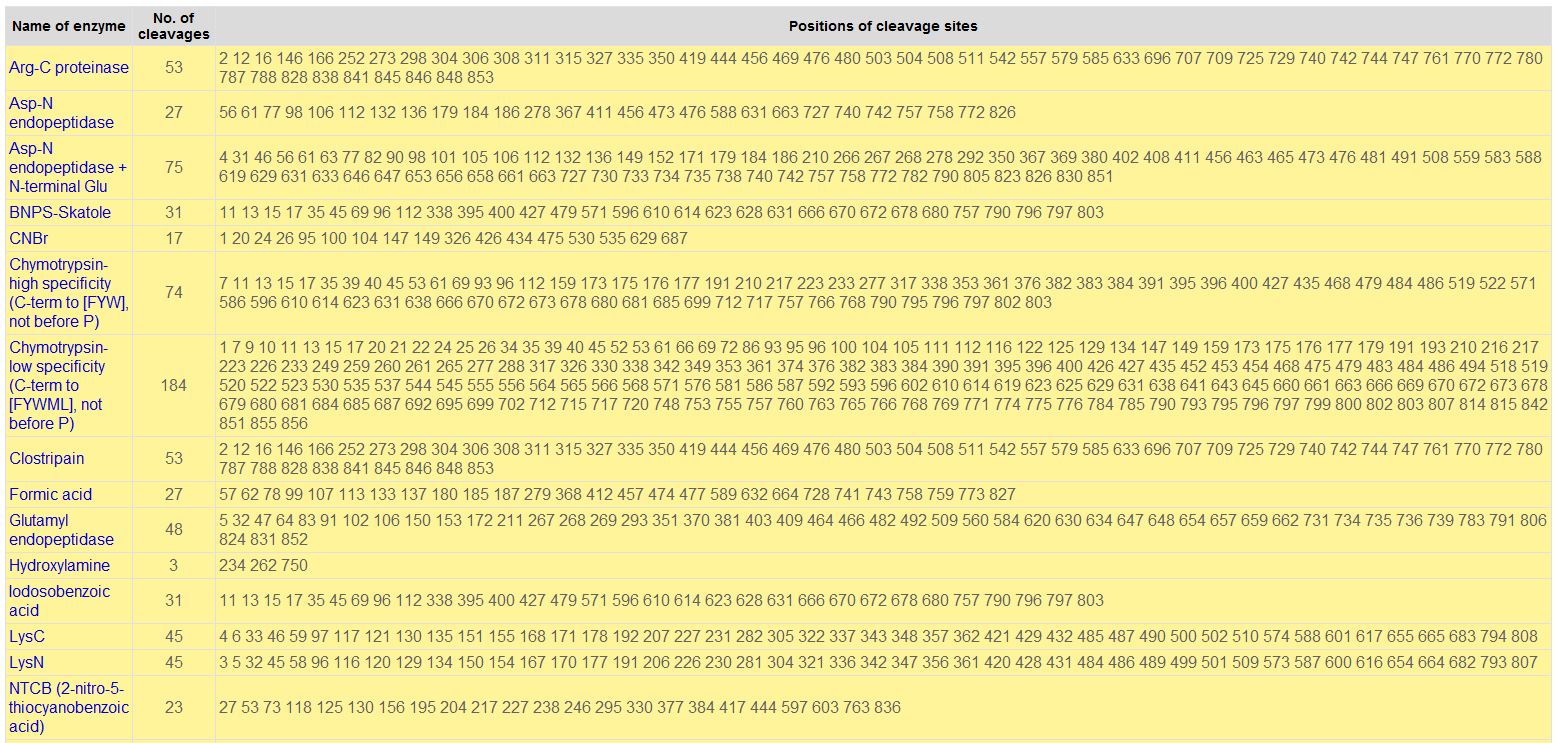
- Digesting a Protein in a Computer:
Sample of Cleavage Results
Doing Primary Structure Analysis:
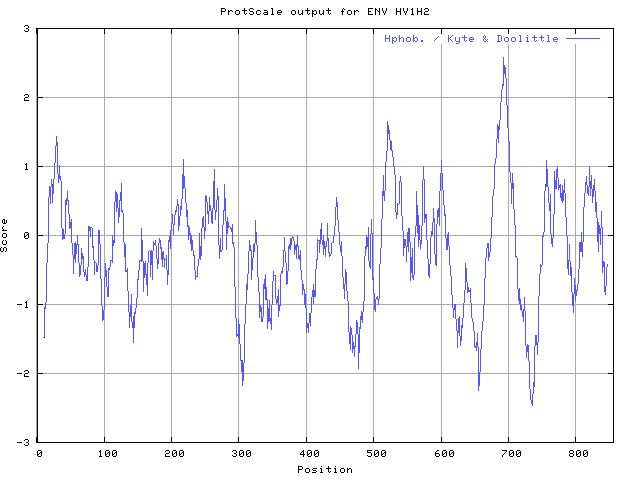
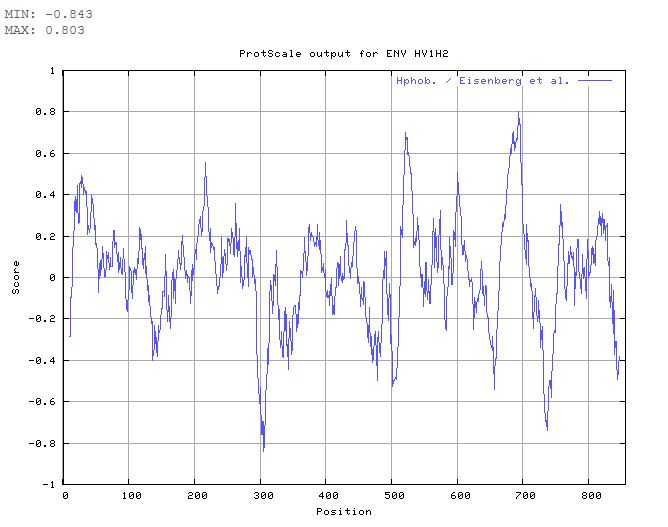
- Looking for Transmembrane Segments-- Running Protscale:
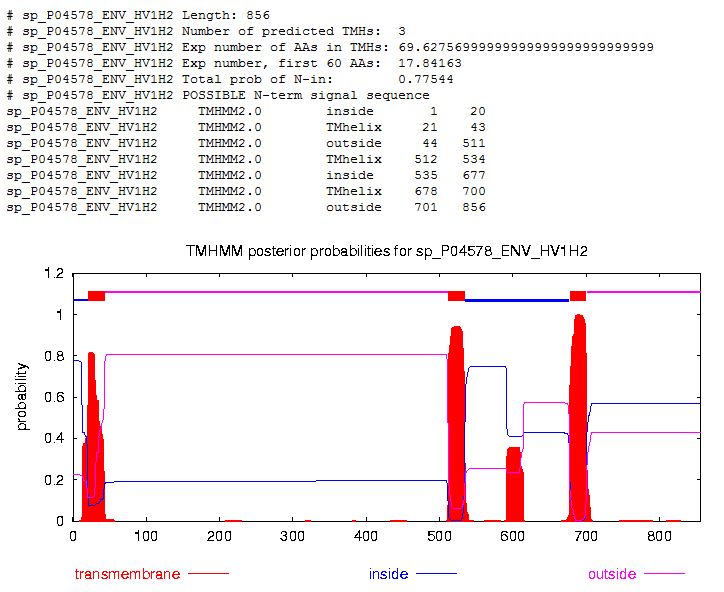
- Looking for Transmembrane Segments-- Running TMHMM:
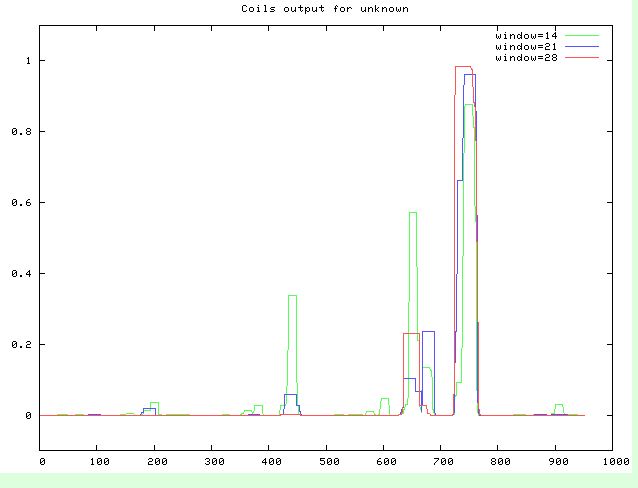
- Looking for coiled-coil regions:
Predicting Post-Translational Modifications in Your Protein:
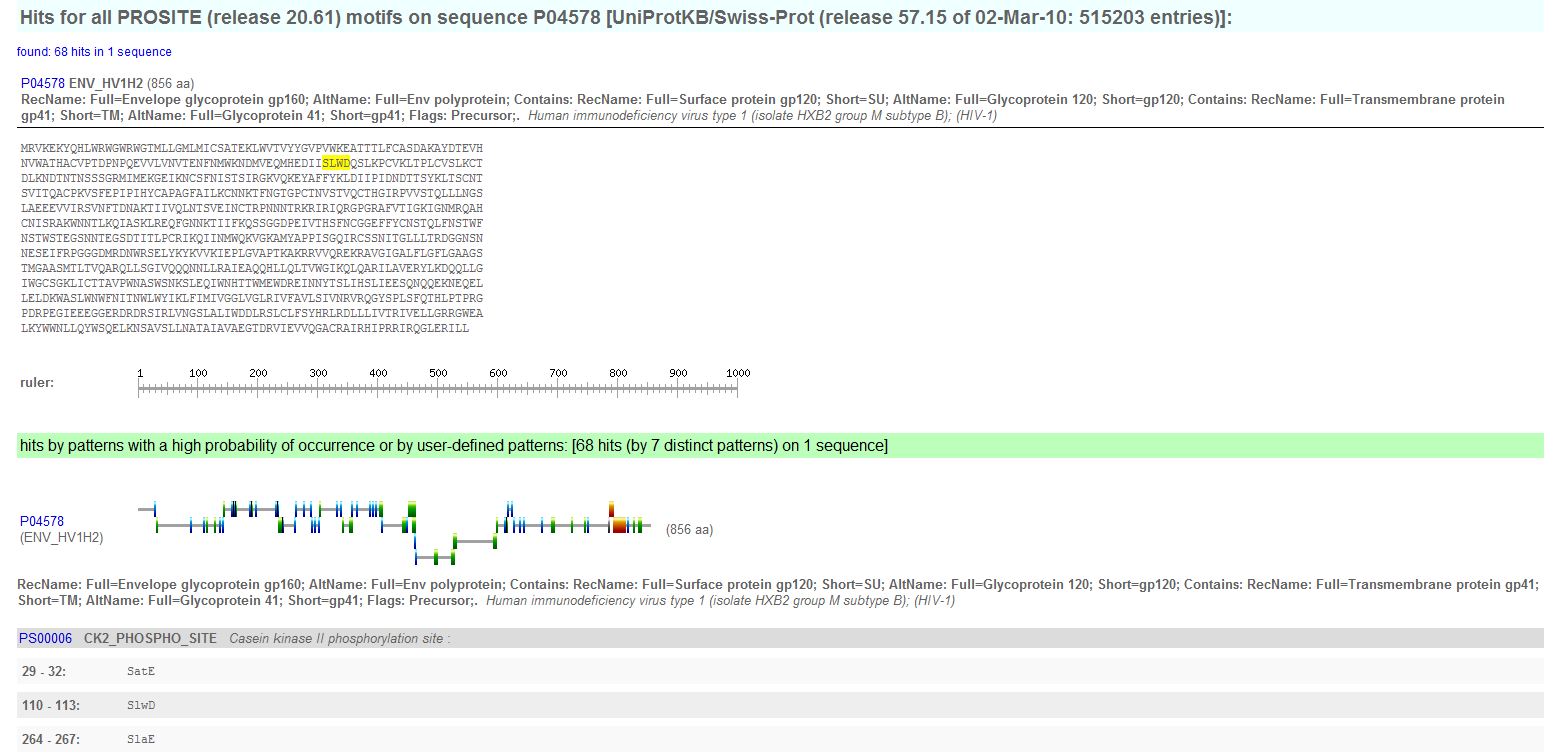
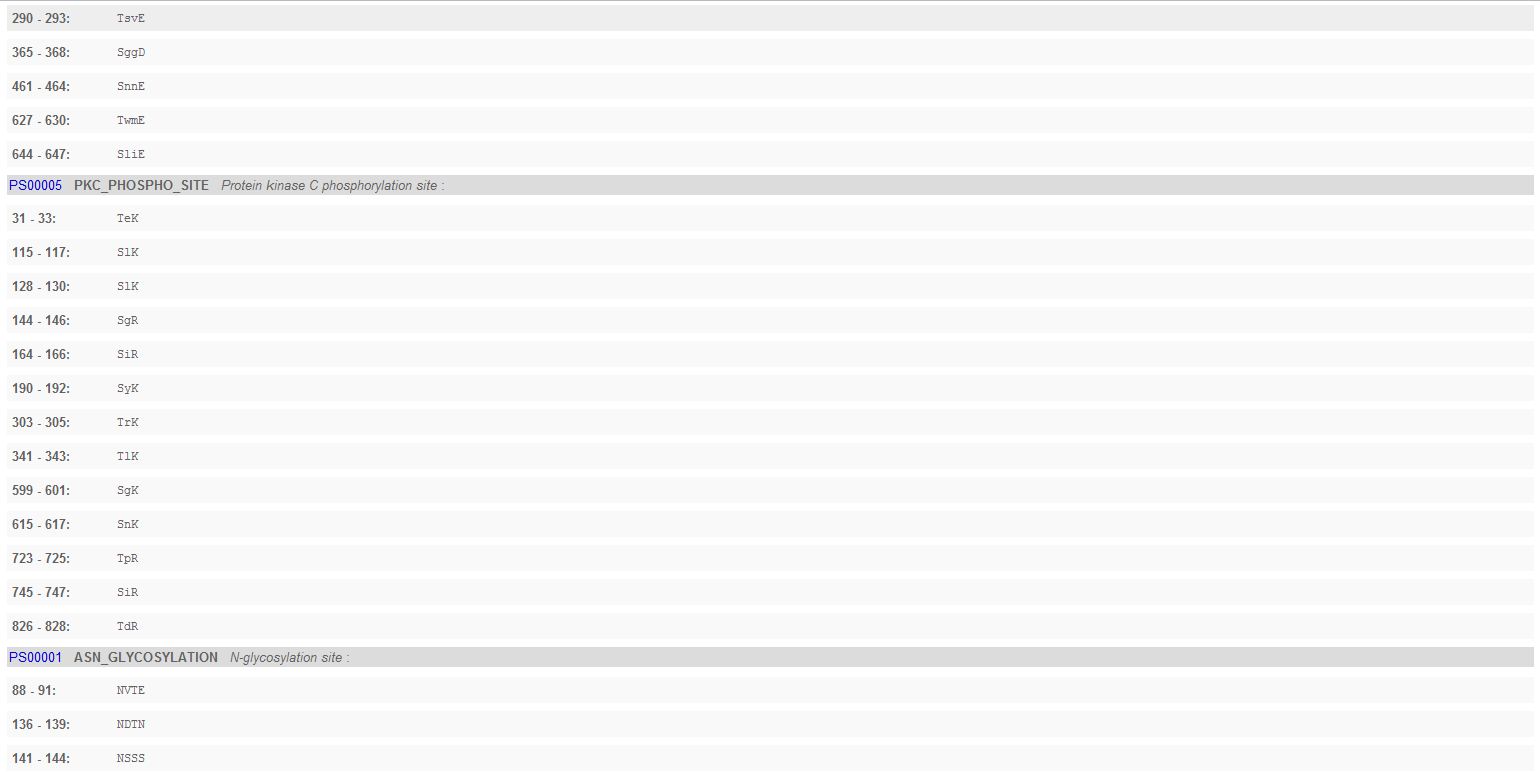
- Looking for PROSITE Patterns:
Finding Known Domains in Your Protein:
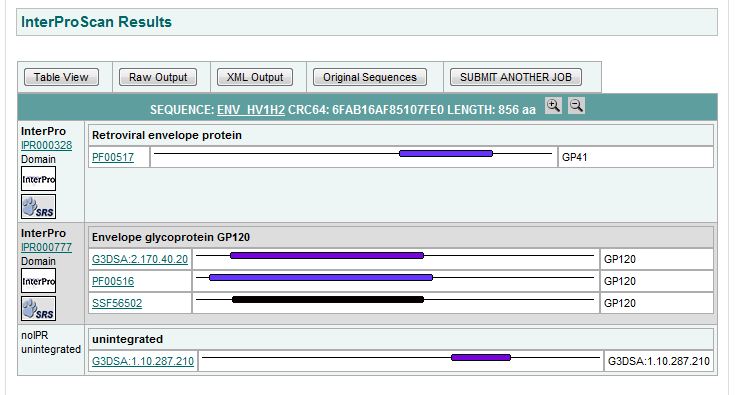
- Choosing the Right Collection of Domains--Finding Domains with InterProScan:
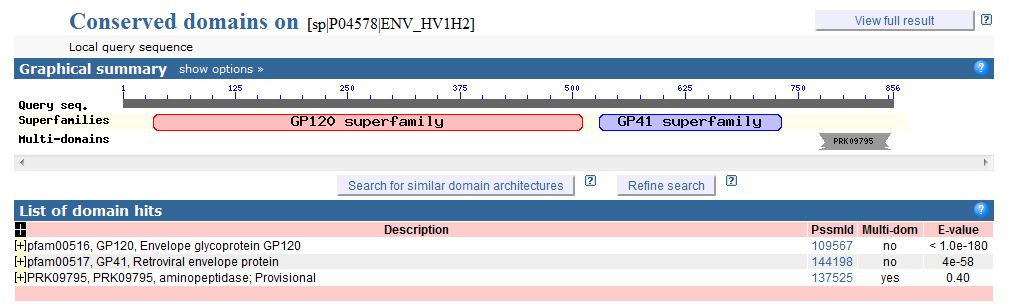
- Choosing the Right Collection of Domains--Finding Domains with CD Server:
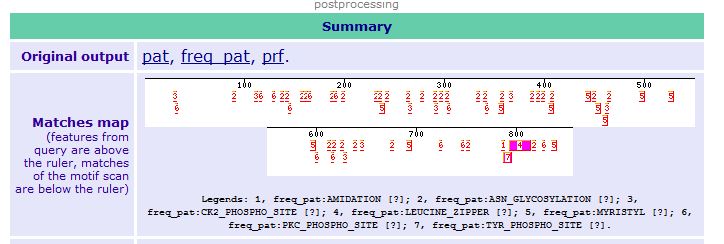
- Choosing the Right Collection of Domains--Finding Domains with Motif Scan:
Class Links
Journal Assignments
- Shared Journal
- BIOL398-01/S10:Class Journal Week 2
- BIOL398-01/S10:Class Journal Week 3
- BIOL398-01/S10:Class Journal Week 4
- BIOL398-01/S10:Class Journal Week 5
- BIOL398-01/S10:Class Journal Week 6
- BIOL398-01/S10:Class Journal Week 7
- BIOL398-01/S10:Class Journal Week 8
- BIOL398-01/S10:Class Journal Week 9
- BIOL398-01/S10:Class Journal Week 11
- BIOL398-01/S10:Class Journal Week 12
- BIOL398-01/S10:Class Journal Week 13
- Assignments