IGEM:Harvard/2006/cyanobacteria/peng labbook
From OpenWetWare
Jump to navigationJump to search
Notebook
Below are notes taken during meetings or during lab; they will be either in pdf or below depending on platform used. Let me know if you need a native OneNote copy instead.
June 12th
Morning
- Notes from first meeting (pdf)
Afternoon
- Explained BioBricks Format
- Created 1% Agarose Gel
- Folding DNA nanostructures
- Transformation of R0010, E7104, E0241 into Top-10 Competent E. Coli
R0010 - Lac operon promoter E7104 - T7 promoter + GFP E0241 - GFP
June 13th
Morning
- Brainstorming by presenting previous projects
- Past_project_notes
Afternoon
- TF's created a 1% Agarose gel which we ran our DNA scaffold samples on
- 130V, 45m+, imaged afterwards
DNA Scaffold types
-both
-scaffold only
-oligos only
- Checked plating of transformant cells; no growth on control
- Grew transformant cells in liquid culture (5mL) overnight with 50uL ampicillin
- More brainstorming - Perry presented his domain swapping experiment, discussed DNA nanostructure box
June 14th
Morning
- DNA Miniprep of transformant colonies
- Out of 5mL of liquid culture, reserved 1mL for gylcerol+freeze and 4mL other
- pelleted E. coli for each sample
- followed QIAprep Miniprep Kit for Microcentrifuge directions
- Used warm dH20 for elution
- Put at 40C for ~2 min to evaporate ethanol before elution
- Forgot to label after elution --> don't know what is what
- Sol'n: During digest will have to run PCR, can tell R0010 from rest, but E7104/E0241 is only different by 30bp; if doesn't work can flip and try two experiments.
- Nanodrop demonstration
Nanodrop results -1: 31.2ng/uL 260/280:1.75 260/230:1.92 -2: 38.5ng/uL 260/280:1.83 260/230:1.87 -3: 33.8ng/uL 260/280:1.70 260/230:1.44
PROBLEM: Messed up labeling of the plasmids! To diagnose, ran a 15min e-Gel to find out which is R0010.
- Digestion of vector/insert
- Digested R0010 (200bp cutout) as vector at S and P site.
- .5uL Spe1, .5uL Pst1
- 11uL h20
- 2.5uL 10X BSA
- 2.5uL #2 NebBuffer
- 8uL DNA
- Digested other 2 (~900bp cutout) as insert at X and P site.
- .5uL Xba1, .5uL Pst1
- 11uL h20
- 2.5uL 10X BSA
- 2.5uL #3 NebBuffer
- 8uL DNA
- Incubate @ 37C for 1h
- Digested R0010 (200bp cutout) as vector at S and P site.
- Phosphatase
- 80C@15min to kill enzyme activity
- Used CIP (1 unit) into the R0010, 1h@37C
- Run on 1% agarose gel
- Image, Cutout, and Purify
- Can isolate the three from the gel
Result
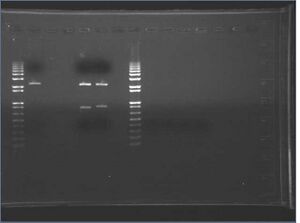
Ladder=1kb+ Lane 1=R0010 (#1) Lane 2=E0241 (#2) Lane 3=E7104 (#3)
June 15th
Morning
- Brainstorming with Pam
- Infeasibility of DNA nanostructures
- (Linked)
- Conduct ligation reaction
- 6uL insert, 2uL vector, 2uL buffer, 10uL other buffer
- 5 min incubation @ RT
- Conduct transformation
- 20mL cells for positive, negative, and exp. ea (top 10)
Afternoon
- Plate transformant cells on LB-CARB
- Brainstorming session in the afternoon
- Linked
June 16th
Morning
- Talk with Prof. Shih about DNA nanostructures and vailidity
- Linked
Afternoon
- Talks on two papers
- Other talks can be found at IGEM:Harvard/2006/Brainstorming
June 17th (Sat)
Afternoon
- Met up in the evening to conduct research on cyanobacteria with Hetmann, Dave, and Jeff
June 18th (Sun)
Afternoon
- Finalized information and created Powerpoint
- IGEM:Harvard/2006/Cyanobacteria for link to cyanobacteria information
- Media: Presentation.ppt for Powerpoint delievered (requires Beta 2007 to work correctly)
June 19th (Week 2)
Morning
- Presentation of four projects
- DNA nanostructures (Matt Katie Valerie Tiffany)
- Fusion Proteins (Perry)
- Universal Cell Signaling (Lewis)
- Cyanobacteria (Peng Hetmann David Jeff)
- Feedback on cyanobacteria
- Found two sources for our plasmids: WHOI and MIT links
- E. coli link may not work, but should figure out synthesis
- Codon Devices can do it for $0.30/bp
- Prof. Alexander van Oudenaarden works with cyanobacteria, [1]
Afternoon
- Prof. Shih's discussion on the honeycomb lattice
- Check plates for GFP expression
Two colonies on experimental (R0010+E0241)! Many on + None on control
- Grow R0010+E0241 in 5mL LB + 50uL Amp (5mg/uL orig) overnight @37
- Further brainstorming
- Emailed Eric Webb about Fedex #; Peter Weigele can give us PCC7942 on Thurs.
June 20th
Morning
- Ordered culture BC11 by Sigma
- Alain ordered PCC7942 and WH8104 strain from WHOI
- Need to look up if other strains grow faster / better to culture
- Need to get ahold of Prof. Knoll and vanO lab to ask about cyanobacteria culture
- Make glycerol stocks of R0010+E0241
- 100uL 50% glycerol, 100uL liquid media
- DNA Miniprep
- Follow handout; had 2 5mL samples, only used one of them
- Tubes 0.5mL PCR; labeled in blue on top
- Digest for diagnosis
- Xba1 and Pst1
- .5uL Xba1, .5uL Pst1
- 11uL h20
- 2.5uL 10X BSA
- 2.5uL #3 NebBuffer
- 8uL DNA
- Incubate @ 37C for 1h
- Xba1 and Pst1
Afternoon
- Research into cyanobacteria: see IGEM:Harvard/2006/Cyanobacteria for contribution
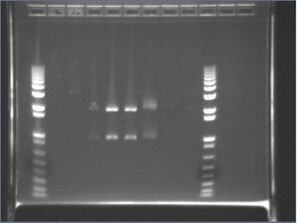
- Digest gel result
June 21st
Morning
- Populating IGEM:Harvard/2006/Cyanobacteria
- Organizing shopping list/calling stores
Afternoon
- Roadtrip to Home Depot to obtain items
- Wrote Guide to building a cyanobacteria incubator
- Built incubator
- Will pick up PCC7942 tomorrow.
June 22nd
Morning
- Working incubator!
- WH8102 came in the mail with SH media
Afternooon
- Went to go pick up PCC7942 and 6803 from Peter Weigele @ MIT Building 68
- Made liquid culture without thiosulfate
- Made 6 samples
- PCC7942 streak w/toothpick
- PCC7942 single w/toothpick
- PCC7942 single w/o toothpick
- PCC6803 streak w/toothpick
- PCC6803 single w/toothpick
- Control w/ toothpick
- Sprayed with EtOH beforehand working area
- 16h day / 8h night, shaking, open lid, 30C
June 23rd
Morning
- Measured light intensity with lux meter
- Turns out only a little region has ~4200 lux; rearranged samples
- Tested plastic cover; does not diffuse light
Afternoon
- General research on ideal conditions for growth
June 26th (Week 3)
Morning
- Morning meetings
- Made more BG11 liquid media w/ thiosulfate
- Made 500x Thiosulfate+Na stock sol'n
- Brought down solid plate recipe
Afternoon
- Checked experiments
- Created new liquid colonies
- 125mL BG11 liquid in autoclaved glass 500mL earlemeyer
- PCC7942 dried out!
- Question to be addressed:
- How do the plasmids work? DNA delivery?
Evening
- Emailed Prof. Susan Golden
June 27th
Morning
- PCR tutorial
- Tm = 65, y = Tm -5
- deltaG > -3 kCal/mol
Afternoon
- Beginning primer design
- Ordered PCC7942 from atcc.org
June 28th
Morning
- Sick :(
Afternoon
- Reviewed information from Prof. Golden
- Growth in PCC6803!
- Completing primer design for KaiABC extraction
July 6th
Morning
- Rehydrated oligos that arrived (10 of them)
- First in 250uL dh20
- Then diluted to 20uM ea
- PCR (colony) from dried PCC7942 plate
- Did not work
- Decided to model oscillation
July 7th
- Recieved PCC7942
- PCRed again, this time with liquid culture as template
- 2 liquid cultures made, 500mL flask w/100 mL
- 1 w/ thiosulfate
- 1 w/o thiosulfate
- 2 solid cultures made
- Modeling
Post July 7th
Look into our cyanobacteria page at IGEM:Harvard/2006/Cyanobacteria/Notebook.