IGEM:Harvard/2006/DNA nanostructures/Notebook/2006-8-16
Repeat PEG Precipitation
Overview
- concentrate on 30 mM MgCl2 folding buffer, 1x oligos trial from yesterday (yielded best PEG results)
- folding rxns:
32 (40 μL rxn) c5.0C (no latch, inside ligand)
32 (40 μL rxn) c5.0D (no latch, outside ligand)
8 (40 μL rxn) c5.0A (no latch, no ligand)
- repeat 8%, 10% PEG precipitation
Folding Reactions
- reconstituted the speedvaced 5.0.C and 5.0.D reactions from yesterday in 200 μL. Essentially we could have skipped the speedvac because as it turns out we're just going to be using the 30mM MgCl2. If we'd needed 20mM we would have reconstitued in 16 μL.
- Made more 10x folding buffer (300 mM MgCl2)
- folding rxns:
32 (40 μL rxn) c5.0C (no latch, inside ligand)
32 (40 μL rxn) c5.0D (no latch, outside ligand)
8 (40 μL rxn) c5.0A (no latch, no ligand)
- Each rxn:
16 ul oligos
9 ul p7308
11 ul H20
4 ul 10x folding buffer
PEG ppt
- protocol: mix all ingredients, add nanostructures last, total volume is 100 mL, incubate on ice for 15 min, spin for 10 min at 16k rcf, pipet off supernatant, resuspend pellet in 100 μL water
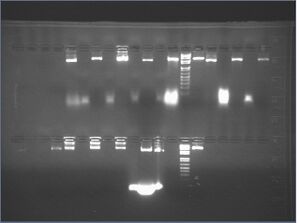
| lane | contents | loading dye |
| 1 | 8 μL untreated 5.0.A | 3 μL |
| 2 | 20 μL 5.0.A in 7% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 3 | 8 μL 5.0.A in 7% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 4 | 20 μL 5.0.A in 8% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 5 | 8 μL 5.0.A in 8% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 6 | 20 μL 5.0.A in 9% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 7 | 8 μL 5.0.A in 9% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 8 | 20 μL 5.0.A in 10% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 9 | 8 μL 5.0.A in 10% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 10 | 1 kb+ ladder | 3 μL |
| 11 | 2.7 μL p7308 | 3 μL |
| 12 | 8 μL untreated 5.0.C | 3 μL |
| 13 | 20 μL 5.0.C in 7% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 14 | 8 μL 5.0.C in 7% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 15 | 20 μL 5.0.C in 8% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 16 | 8 μL 5.0.C in 8% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 17 | 20 μL 5.0.C in 9% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 18 | 8 μL 5.0.C in 9% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 19 | 20 μL 5.0.C in 10% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 20 | 8 μL 5.0.C in 10% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 21 | 8 μL untreated 5.0.D | 3 μL |
| 22 | 20 μL 5.0.D in 7% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 23 | 8 μL 5.0.D in 7% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 24 | 20 μL 5.0.D in 8% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 25 | 8 μL 5.0.D in 8% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 26 | 20 μL 5.0.D in 9% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 27 | 8 μL 5.0.D in 9% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 28 | 20 μL 5.0.D in 10% PEG and 0.5 M NaCl (supernatant) | 3 μL |
| 29 | 8 μL 5.0.D in 10% PEG and 0.5 M NaCl (pellet) | 3 μL |
| 30 | 1 kb+ ladder | 3 μL |
| 31 | 2.7 μL p7308 | 3 μL |
| 32-40 | (empty) | 3 μL |
Protection assay
- used nanostructures folded above that were purified with 8% PEG, 0.5 M NaCl
- mixed the following reactions, adding enzyme last
- attachment DNA used: c4.0.6.1ob (37 bp)
- oligo-ligand: script output (45 bp)
- incubated at 37[[:Category:{{{1}}}|{{{1}}}]] for 60 min.
- 10 μL each reaction (with 2 μL loading dye added) run on 18% Tris-Gly PA gel for 100 min. at 120 V, stained with SYBR Gold, imaged under EtBr filter
- other 10 μL stored at 4[[:Category:{{{1}}}|{{{1}}}]] overnight, then added 2 μL loading dye to each, and ran on 4%-20% gradient TBE PA gel for 110 min at 120 V, stained with SYBR Gold, imaged under EtBr filter
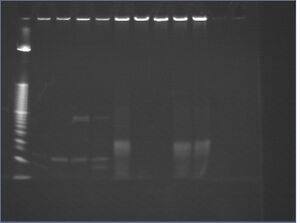
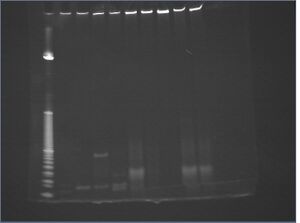
| lane | trial | DNA | 10x NEBuffer 4 | 10x BSA | AscI | water |
| 1 | 10 bp+ ladder | 5 μL | ||||
| 2 | -nanostructures -ligand | 1 μL 1 μM attachment DNA | 2 μL | 2 μL | 1 μL 500 U/mL | 14 μL |
| 3 | -nanostructures -attachment | 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 1 μL 500 U/mL | 14 μL |
| 4 | -nanostructures -enzyme | 1 μL 1 μM attachment DNA, 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 0 μL | 14 μL |
| 5 | -nanostructures | 1 μL 1 μM attachment DNA, 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 1 μL 500 U/mL | 13 μL |
| 6 | inward-facing ligands | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.C | 2 μL | 2 μL | 1 μL 500 U/mL | 5 μL |
| 7 | outward-facing ligands | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.D | 2 μL | 2 μL | 1 μL 500 U/mL | 5 μL |
| 8 | -oligos | 2.25 μL 44 nM p7308 | 2 μL | 2 μL | 1 μL 500 U/mL | 13.75 μL |
| 9 | -enzyme | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.C | 2 μL | 2 μL | 0 μL | 6 μL |
| 10 | -ligand | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.A | 2 μL | 2 μL | 1 μL 500 U/mL | 5 μL |
results/discussion
- lane 2: attachment DNA shows two bands (at 20 bp and 30 bp)
- unusual, but not fatal
- lane 3: oligo-ligand shows at 30 bp
- lane 4: ds construct shows at 85 bp, excess oligo-ligand at 30 bp
- lane 5: digested construct shows at 15 bp, undigested traces at 85 bp, excess oligo ligand at 30 bp
- digest on ds construct (postive control) is successful
- lanes 6 and 9: no visible difference between nanostructures when enzyme is added (lane 6) and when it is not added (lane 9)
- implies that oligo streaks are unremoved oligos
- expected because both have inward-facing oligos
- lanes 6 and 10: no visible difference between nanostructures when oligo-ligand was folded in (lane 6) and when it was not (lane 10)
- again, implies that oligo streaks are unremoved oligos
- expected because neither has outward-facing oligos
- lane 7: most oligos appear to be removed
- lane 8: scaffold is not digested
- thank goodness
- N.B. -- we originally thought that C had outward facing ligands and D had inward facing ligands, not vice versa. thus, the experimental design was, uh, flawed. we'll try this again properly.
- gradient TBE
- even lane 7 (D) shows some oligo contamination
- lane 5 shows poor resolution between digested DNA and main digest products
conclusions
- perhaps need stronger digest conditions (more enzyme?) and more nanostructures
- purer samples are needed, but this is not impossible (lane 7 looks great)
- repeat PEG precipitation on resuspended samples?
- gradient TBE better for detecting oligo traces, but not as good for visualizing digest product
Protection assay again
- change: doubled enzyme amount, doubled amount of product loaded into gel
- change: used 7% PEG-ppt'd 5.0.A and 5.0.C, maybe they'll be purer
- did not change: gel buffer, which could explain poorer gel quality
- used nanostructures folded above that were purified with 7% (A, C) or 8% (D) PEG, 0.5 M NaCl
- mixed the following reactions, adding enzyme last
- attachment DNA used: c4.0.6.1ob (37 bp)
- oligo-ligand: script output (45 bp)
- incubated at 37[[:Category:{{{1}}}|{{{1}}}]] for 60 min.
- 20 μL each reaction (with 2 μL loading dye added) run on 18% Tris-Gly PA gel for 110 min. at 120 V, stained with SYBR Gold, imaged under EtBr filter
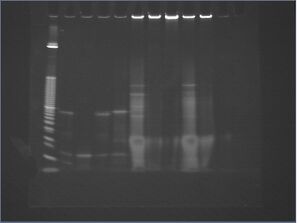
| lane | trial | DNA | 10x NEBuffer 4 | 10x BSA | AscI | water |
| 1 | 10 bp+ ladder | 5 μL | ||||
| 2 | -nanostructures -ligand | 1 μL 1 μM attachment DNA | 2 μL | 2 μL | 2 μL 500 U/mL | 13 μL |
| 3 | -nanostructures -attachment | 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 2 μL 500 U/mL | 13 μL |
| 4 | -nanostructures -enzyme | 1 μL 1 μM attachment DNA, 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 0 μL | 14 μL |
| 5 | -nanostructures | 1 μL 1 μM attachment DNA, 1 μL 1 μM oligo-ligand | 2 μL | 2 μL | 2 μL 500 U/mL | 12 μL |
| 6 | outward-facing ligands | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.C | 2 μL | 2 μL | 2 μL 500 U/mL | 4 μL |
| 7 | inward-facing ligands | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.D | 2 μL | 2 μL | 2 μL 500 U/mL | 4 μL |
| 8 | -oligos | 2.25 μL 44 nM p7308 | 2 μL | 2 μL | 2 μL 500 U/mL | 11.75 μL |
| 9 | -enzyme | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.C | 2 μL | 2 μL | 0 μL | 6 μL |
| 10 | -ligand | 10 μL 10 nM 8%-PEG-purified, ligand-incubated c5.0.A | 2 μL | 2 μL | 2 μL 500 U/mL | 4 μL |
results/discussion
- postive control (lane 5) does not show a clear digest, so we really can't conclude much about this gel
- also: use fresh buffer, because this looks ugly
- N.B. -- again, we still thought that C had outward facing ligands and D had inward facing ligands, not vice versa. thus, the experimental design was, uh, flawed. we'll try this again properly.
Magnetic Streptavidin Protection, Take 2
- Steps 9-11 were done from the Tues 8.15 protocol.
- Mistakes:
- The Speedvac was allowed to proceed for an hour. Upon return, what had been thought to be 200uL of final-supernatant had completely dried - unless the tubes that had been speedvaced were not in fact the tubes that had contained the final-supernatant, but intermediate tubes. Because the initial final-supernatant had appeared slightly brown, these tubes were placed back into the MagnaRack and the supernatants were re-extracted into different, final tubes. These tubes might have accidentally been thrown out in the cleaning of the lab area that occured while I was out to lunch. If so, there might be some small residual amount of elute which might show up in the gels, but is unlikely to. If this is the case, the experiment will need to be repeated (cost of beads: ~$32).
- If the tubes were the correct ones, the fact that they were Speedvaced to drying might have damaged the nanostructures irreparably and made them difficult, if not impossible to visualize in the gel. But hopefully scaffold and oligos will still be able to be seen.
- Gel:
| Lane | Component | Amount |
| 1 | 1kb ladder | 10uL |
| 2 | p7308 | 9uL |
| 3 | 6hb wash-supernatant | 30uL |
| 4 | c5.0.8b wash-supernatant | 30uL |
| 5 | c5.0.Eb wash-supernatant | 30uL |
| 6 | c5.0.Fb wash-supernatant | 30uL |
| 7 | 6hb final-supernatant (?) | 30uL |
| 8 | c5.0.8b final-supernatant (?) | 30uL |
| 9 | c5.0.Eb final-supernatant (?) | 30uL |
| 10 | c5.0.Fb final-supernatant (?) | 30uL |
- Results:
- As would concur with the suspicion that the wrong tubes had been Speedvaced, none of the four lanes of final-supernatant show any elute at all. The fact that there is no band in the biotinylated oligo final-supernatant lane (ie. lane 8) AND none in its wash-supernatant lane (ie. lane 4) suggests that the the biotinylated oligos were all bound by the streptavidin and then eluted off - but not into the "final-supernatant" tube. Thus, the experiment needs to be reconducted.
- The bright bands that seem to be trapped in the wells of lanes 3 and 5 (ie. 6hb wash and Eb wash) suggest that some damage was suffered by the nanoboxes during the Speedvacing. There are two fainter bands in each of these lanes, one running slightly higher (slower) than the p7308, one running lower (faster) than the p7308. This could be undamaged nanostructure - however, since no c5.0.A (c5.0 barrel) was run as a control, no definite conclusions can be drawn.
- The fact that lane 5 (Eb wash) shows such a large amount of nanoboxes, whereas lane 6 (Fb wash) does not, implies that Eb ("outside" biotinylation) was not bound by the streptavidin while Fb ("inside" biotinylation) was, as was true in the last gel (previously thought to be due to a mix up of the tubes). The most likely conclusion to be drawn is that there was mix up in the creation of the working stocks or pre-working stocks: Eb must truly be biotinylated on the inside, Fb on the outside, which may be due to a mistake in mixing c5.0.8b instead of c5.0.9b into Fb and vice versa, or because the oligos intended for the c5.0.8b pre-working stock were confused with those for c5.0.9b and vice versa.
- Upon further investigation:
- The sequences for c5.0.8.1 and c5.0.9.1 on the tube match those on the final oligo list for c5.0.
- Oligo 7 (pre-split c5.0.8.1) correctly has an OUT aptamer site, and oligo 33 (pre-split c5.0.9.1) correctly has an IN aptamer site.
- The best conclusion is that the pre-working or working stocks were incorrectly mixed.
- Either way, correct working stocks and correct pre-working stocks must be made up and the former ones thrown out.
- Upon further investigation:
- The fact that lane 5 (Eb wash) shows such a large amount of nanoboxes, whereas lane 6 (Fb wash) does not, implies that Eb ("outside" biotinylation) was not bound by the streptavidin while Fb ("inside" biotinylation) was, as was true in the last gel (previously thought to be due to a mix up of the tubes). The most likely conclusion to be drawn is that there was mix up in the creation of the working stocks or pre-working stocks: Eb must truly be biotinylated on the inside, Fb on the outside, which may be due to a mistake in mixing c5.0.8b instead of c5.0.9b into Fb and vice versa, or because the oligos intended for the c5.0.8b pre-working stock were confused with those for c5.0.9b and vice versa.
- The bright bands that seem to be trapped in the wells of lanes 3 and 5 (ie. 6hb wash and Eb wash) suggest that some damage was suffered by the nanoboxes during the Speedvacing. There are two fainter bands in each of these lanes, one running slightly higher (slower) than the p7308, one running lower (faster) than the p7308. This could be undamaged nanostructure - however, since no c5.0.A (c5.0 barrel) was run as a control, no definite conclusions can be drawn.
- There are no bands in lane 6 (Fb) other than one that runs at the same speed as p7308, and this considerably fainter than the total brightness in lane 5 (Eb), suggesting that the nanoboxes have been bound by streptavidin.
This is a positive indicator that the protection assay worked, as are the other results from this gel, but to be certain, the experiment must be run again with the final-supernatants present.
Magnetic Streptavidin Protection, Take 3
(See above) The best thing to do would be to make up new tubes of pre-working stocks c.5.0.8(b) and 9(b), and working stocks c.5.0.E(b) and F(b). However, at this time the oligos required to make more c5.0.1, which we've run out of, are at the Medical School. Thus, the next best thing was to use the two remaining folded reactions of E(b) and F(b), which are really F(b) and E(b) respectively, and repeat the experiment.
(Generally the same protocol was used as previously, with a few modifications, which are in red.)
- Protocol:
- 1. Incubate:
- 200uL beads
- 160uL 1x folding buffer
- 40uL test solution
- 1. Incubate:
- TEST SOLUTIONS:
- c5.0.D (to show that a plain unbiotinylated structure won't bind streptavidin, and for a control that runs at the same rate in a gel as the E(b) and F(b))
- 10uL of biotinylated oligos (c5.0.8(b)) + 30uL H2O (=500pmoles, comparable to reactions' binding capacity)
- c5.0 E(b) (outside biotinylation) - unpurified, because was folded at 10mM MgCl2 and an appropriate PEG precipitation percentage had not been determined for this folding condition
- c5.0 F(b) (inside biotinylation)
- TEST SOLUTIONS:
- 2. Mix by thorough trituration
- 3. Pellet by drawing magnet down to bottom of tube (~ 1 minute using the MagnaRack)
- 4. Remove wash-supernatant to separate tube (all supernatant from these washes were collected in one tube per reaction)
- 5. Add 200uL 1x folding buffer
- 6. Repeat steps 2-5 three more times, placing the supernatant each time into the corresponding wash-supernatant tube.
- 7. Trypsinize by adding to pellet:
- 5uL trypsin (1mg/mL)
- 195uL 1x folding buffer
- 8. Incubate overnight @ 37 degrees C
- 7. Trypsinize by adding to pellet:
- 9. Simultaneously take the wash-supernatant tubes and pellet any residual beads using the MagnaRack. Transfer the supernatant to new, clean tubes; discard the old tubes containing the residual beads.
- 10. Pellet (1 minute using the MagnaRack) and remove the final-supernatant to a clean tube. Retain the pellet at 4 degC, in case further trypsinization is determined to be needed after the gel is run.
- 11. Speedvac down, until the final-supernatants are dry (1hr 30min @ 30C). :
- the 4 wash-supernatants for each of the test solutions
- the 4 final-supernatants for each of the test solutions
- Reconstitute the final-supernatants in 30uL 1x folding buffer.
- 12. Run 30uL of each supernatant on 2% agarose gel (10mM MgCl2) for 1hr at 80V.
- Steps 9-11 were done Th 8.17.
