IGEM:Harvard/2006/DNA nanostructures/Notebook/2006-8-11
From OpenWetWare
Jump to navigationJump to search
Thoughts/ramblings/goals/questions/general frustrations
The results of yesterday's experiment show that Microcon filtration gives low yields and the PEG precipitation (at least at 10%) damages nanostructures regardless of folding conditions.
Questions
- Are Microcon yields unacceptably low, or are they acceptable? (Can we use the NanoDrop to quantify our yield?)
- Gels are much better for quantifying yield - you can try both and see how they compare.
- Low concentrations of PEG should precipitate large nanostructures. Will some smaller concentration of PEG not harm nanostructures formed under some folding conditions?
- Our August 2 experiment showed that even low concentrations of PEG damage nanostructures folded under "standard" conditions (10x oligos, 10 mM MgCl2).
Gigundo PEG precipitation
- goal: test 0% to 8% PEG precipitations with nanostructures folded under all six folding conditions from yesterday
- optimistic hypothesis: nanostructures folded with higher concentrations of oligos and/or MgCl2 will show less damage after treatment with PEG
Protocol: prepare the following 30 samples.
| Trial | Final PEG % | Gel | Lanes | 20% PEG (μL) | 5 M NaCl (μL) | Nanostructures (μL) | water (μL) | Total volume (μL) |
| 1-0 | 0% | 1 | 1 | 0 | 5 | 5 | 20 | 50 |
| 1-2 | 2% | 1 | 3 | 5 | 5 | 5 | 15 | 50 |
| 1-4 | 4% | 1 | 5 | 10 | 5 | 5 | 10 | 50 |
| 1-6 | 6% | 1 | 7 | 15 | 5 | 5 | 5 | 50 |
| 1 kb+ ladder | 1 | 9 | 10 | |||||
| p7308 | 1 | 10 | 10 | |||||
| 1-8 | 8% | 1 | 11 | 20 | 5 | 5 | 0 | 50 |
| 2-0 | 0% | 1 | 13 | 0 | 5 | 5 | 20 | 50 |
| 2-2 | 2% | 1 | 15 | 5 | 5 | 5 | 15 | 50 |
| 2-4 | 4% | 1 | 17 | 10 | 5 | 5 | 10 | 50 |
| 2-6 | 6% | 1 | 19 | 15 | 5 | 5 | 5 | 50 |
| 2-8 | 8% | 1 | 21 | 20 | 5 | 5 | 0 | 50 |
| 3-0 | 0% | 1 | 23 | 0 | 5 | 5 | 20 | 50 |
| 3-2 | 2% | 1 | 25 | 5 | 5 | 5 | 15 | 50 |
| 3-4 | 4% | 1 | 27 | 10 | 5 | 5 | 10 | 50 |
| 1 kb+ ladder | 1 | 29 | 10 | |||||
| p7308 | 1 | 30 | 10 | |||||
| 3-6 | 6% | 1 | 31 | 15 | 5 | 5 | 5 | 50 |
| 3-8 | 8% | 1 | 33 | 20 | 5 | 5 | 0 | 50 |
| 4-0 | 0% | 1 | 35 | 0 | 5 | 5 | 20 | 50 |
| 4-2 | 2% | 1 | 37 | 5 | 5 | 5 | 15 | 50 |
| 4-4 | 4% | 1 | 39 | 10 | 5 | 5 | 10 | 50 |
| 4-6 | 6% | 2 | 1 | 15 | 5 | 5 | 5 | 50 |
| 4-8 | 8% | 2 | 3 | 20 | 5 | 5 | 0 | 50 |
| 5-0 | 0% | 2 | 5 | 0 | 5 | 5 | 20 | 50 |
| 5-2 | 2% | 2 | 7 | 5 | 5 | 5 | 15 | 50 |
| 1 kb+ ladder | 1 | 29 | 10 | |||||
| p7308 | 1 | 30 | 10 | |||||
| 5-4 | 4% | 2 | 11 | 10 | 5 | 5 | 10 | 50 |
| 5-6 | 6% | 2 | 13 | 15 | 5 | 5 | 5 | 50 |
| 5-8 | 8% | 2 | 15 | 20 | 5 | 5 | 0 | 50 |
| 6-0 | 0% | 2 | 17 | 0 | 5 | 5 | 20 | 50 |
| 6-2 | 2% | 2 | 19 | 5 | 5 | 5 | 15 | 50 |
| 6-4 | 4% | 2 | 21 | 10 | 5 | 5 | 10 | 50 |
| 6-6 | 6% | 2 | 23 | 15 | 5 | 5 | 5 | 50 |
| 6-8 | 8% | 2 | 25 | 20 | 5 | 5 | 0 | 50 |
| 1 kb+ ladder | 1 | 29 | 10 | |||||
| p7308 | 1 | 30 | 10 |
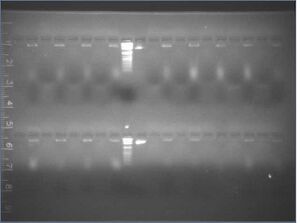
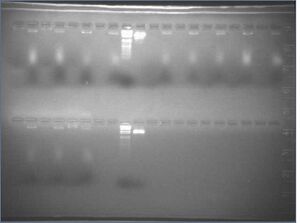
- incubate on ice for 15 min.
- spin at 16 k rcf at 4[[:Category:{{{1}}}|{{{1}}}]] for 10 min.
- carefully pipet off supernatant
- resuspend "pellet" in 20 μL of respective folding buffer
- load resuspended pellets in odd-numbered lanes of 2% TBE agarose gel supplemented to 10 mM MgCl2
- load 30 μL (of 50 μL) of supernatant into adjacent even-numbered lanes (e.g., trial 1-0 has pellet in lane 1 and supernatant in lane 2)
- run at 60V for 1 h
Results/discussion
- PEG precipitations appeared to have failed: no oligos were separated
- curiously, the dye in the "supernatant" lanes ran at about 2/3 of the speed of the dye in the "pellet" lanes, and it gave a smear and not a band
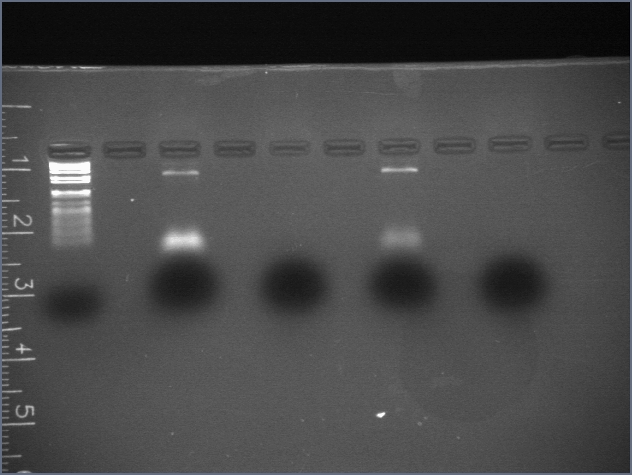
Incubation of 3.2.E with thrombin beads
- Goal: test if we can detect the binding of a nanostructure with outside aptamers to thrombin beads.
100 uL 10 nM 3.2.E or 100 uL 10 nM mix of 6.4. H/I was incubated with 250 uL thrombin beads (supplied as a 50% slurry). 3.2.E has outside aptamer sequences while 6.4.H/I do not. Following a 30 minute incubation, the beads were washed. The beads were then eluted by incubating with 250 uL 50% w/v free thrombin for 30 minutes. Washes and elutions were run on a 2% agarose gel for 60 minutes at 60V.
| Lane | Contents |
| 0 | 1kb+ DNA ladder |
| 2 | 6.4. H/I wash |
| 4 | 6.4. H/I elution |
| 6 | 3.2.E wash |
| 8 | 3.2.E elution |