IGEM:Harvard/2006/DNA nanostructures/Notebook/2006-7-26
Gel purification
gel purified nanoboxes design 3.2.D and 3.2.E (lanes 3 and 4) - not great yield
Goals and questions
Goals
- continue to evaluate the best way to purify nanostructures away from oligos
- run gel of gel purification products
- titrate PEG concentrations
- get streptavidin to stain!
Questions
- can streptavidin stain with silver stain (and so we're doing something wrong), or do we need to find a new stain?
- can we implement an EcoRI restriction of DNA cleavage instead?
- does the p7308 scaffold have any EcoRI restriction sites?
- how long should an oligo be?
- under what conditions can we visualize a short ss oligo?
- SYBR Gold
- adding a complimentary DNA
Oligo design
An ideal box ligand would be a substance that reliably binds to a nanostructure and is easy to detect. Thrombin turned out not to be an ideal ligand because it doesn't reliably bind to a thrombin apatamer, especially under gel conditions. Streptavidin turned out not to be an ideal ligand because although its binding with biotinylated oligos is reliable (or at least we have no reason to assume it's not), streptavidin images poorly with silver staining.
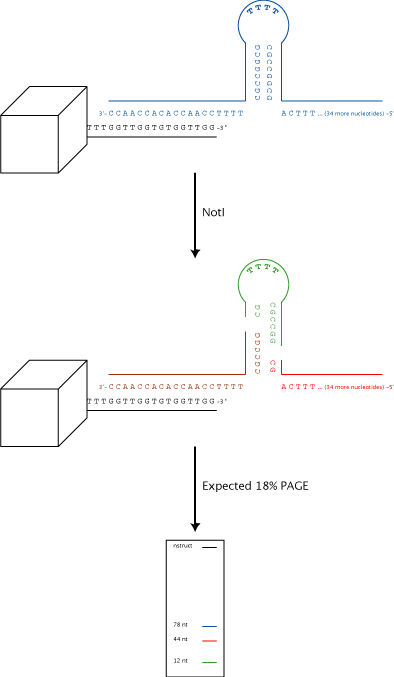
We've decided to try another ligand: a short ssDNA oligo. The 3' end of the oligo would have a 15-bp stretch complimentary to a thrombin aptamer (so that we can use the oligos we already have) and would contain a hairpin with a restriction site. The nanostructure could be incubated with the ssDNA after initial folding, and then adding the appropriate restriction enzyme would cleave a ssDNA that is shorter than the original oligo and should be detectable by PAGE with SYBR Gold staining.
Here's a schematic of the process:
Restriction enzyme choice
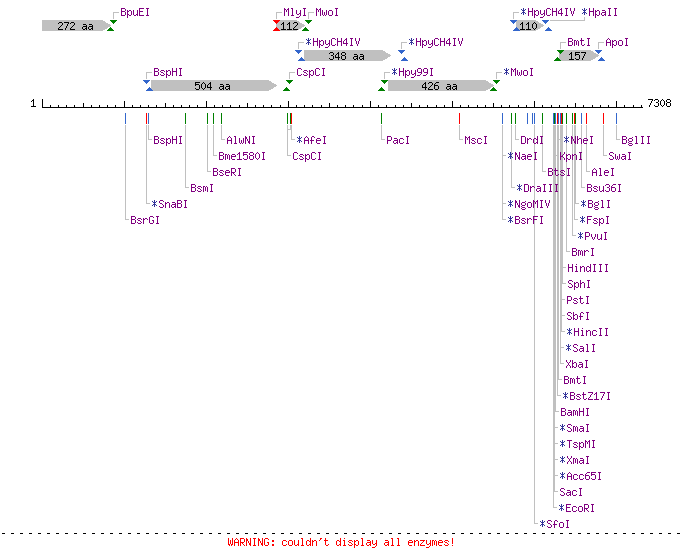
In designing the oligo, we must choose a restriction site that does not appear in the p7308 scaffold, or else the addition of the restriction enzyme would begin to digest our nanostructure. NEB Cutter shows that p7308 has a large multiple cloning site near its 3' end.
A search of the p7308 sequence shows that there is no NotI restriction site (gcggccgc), so we will design our oligo with this sequence.
Random generation script
In accordance with the design above, this code generates 39 random nucleotides (free of NotI sites), appends an NotI site, appends four thymine nucleotides, appends another NotI site, appends four more thymine nucleotides, then appends the reverse compliment of the thrombin aptamer. It was adapted from random sequence generator and restriction site checker.
One program output:
ATGCTGAGGGTGAGCCCCTACTCGTCGTCAACAATTTCAGCGGCCGCTTTTGCGGCCGCTTTTCCAACCACACCAACC
The code:
#!/usr/bin/python
import random
import sys
import string
BamHI = 'ggatcc'
EcoRI = 'gaattc'
NotI = 'gcggccgc'
aptamer = 'GGTTGGTGTGGTTGG'
def rev(s):
return s[::-1]
complement = string.maketrans('ACGTacgt','TGCAtgca')
def comp(s):
return rev(s.translate(complement))
def hasresite(s):
result = False
if s.count(BamHI) > 0:
print 'BamHI found'
result = True
elif s.count(EcoRI) > 0:
print 'EcoRI found'
result = True
elif s.count(NotI) > 0:
print 'NotI found'
result = True
return result
# prints random sequence, length specified as argument
def randseq(l):
flag = True
s = []
while (flag == True):
s = []
for i in range(l):
s.append(random.choice(['a', 'c', 'g', 't']))
flag = hasresite(''.join(s))
return ''.join(s)
#if len(sys.argv) > 1:
# a = int(sys.argv[1])
#else:
# sys.exit("usage: ./random-sequence.py [length]")
final_seq = []
final_seq.append(randseq(39))
final_seq.append(NotI)
final_seq.append('tttt')
final_seq.append(NotI)
final_seq.append('tttt')
final_seq.append(comp(aptamer))
final_seq = ''.join(final_seq).upper()
print final_seq