IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-8-7
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Note: a backslash '\' followed by a pair of letters indicates a digest, e.g. \X-P means an XbaI-PstI digest.
T-shirt design

To-do
Miniprep the 15 ligation transformants we grew up yesterday (10 KaiA+bkb, 5 KaiB+bkb)
LacIq and J04450 (RFP insert)
In today's meeting, George Church recommended putting lacIq on the plasmid.
Gel purification of J04450, high-copy vector, and pSB4A3 low-copy vector
We gel-purified the J04450, high-copy vector, and psB4A3 low-copy vector that we extracted from yesterday's gel.
Miniprep of 15 samples from the KaiA+bkb and KaiB+bkb transformation
15 samples (KaiA+bkb 2-11, KaiB+bkb 2-6) miniprepped from 8uL of culture; made glycerol stock also. Both are sitting in their seperate containers in the freezer or -80 respectively. Nanodrop values varied from 10-20ng/uL - next time only grow in 2uL LB?
Digest X-P to test ligation
KaiA+bkb 1-11 and KaiB+bkb 1-6 were digested with X&P to see if the ligation worked or failed. The protocol based loosely on the Silver protocol, but assuming each sample has ~15ng/uL DNA, is:
- 19.25 uL DNA (~15 ng/uL gives 0.288ug DNA)
- 2 uL H2O
- 2.5 uL buffer (EcoRI buffer or Buffer 3)
- 0.5 uL enzyme 1 (XbaI)
- 0.5 uL enzyme 2 (PstI)
- 0.25 uL BSA
25 uL total volume.
MM:
- 19x2=38 dh20
- 19x2.5= 47.5 buffer 3
- 19x0.5 = 9.5 XbaI
- 19x0.5 = 9.5 PstI
- 19x0.25 = 4.75 BSA
pipet 5.75 into each.
Ligation of J04450 + pSB4A3
We iigated the gel-purified J04450 (\X-P) and pSB4A3 (\E-S) using the Roche rapid ligation kit.
Reaction:
- 2 uL pSB4A3
- 4 uL J04450
- 2 uL DNA dilution buffer (vial 2)
- 2 uL H2O
- 10 uL DNA ligation buffer (vial 1)
- 1 uL ligase (vial 3)
Transformation of J04450 + pSB4A3 into Top10 and Top10F'
We transformed the J04450 + pSB4A3 ligation into Top10 and Top10F' cells, to test for constitutive expression of RFP.
Transformations:
- 30 uL Top10 competent cells, 3 uL ligation
- 30 uL Top10F' competent cells, 3 uL ligation
- 10 uL Top10 competent cells, 1 uL positive control
- 10 uL Top10F' competent cells, 1 uL positive control
- 10 uL Top10 competent cells, 3 uL H2O
- 10 uL Top10F' competent cells, 3 uL H2O
Plated on 8 LB-carb plates: 1 for each of the controls, and 2 for each of the ligation transformations.
Digests
The digests that were completed earlier in PCR machines 1 and 2 (from earlier) were run for 2 hours with a 20 minute heat shock to inactivate the restriction enzymes.
The digests were then run on 1% agarose gel that contained 20 lanes (25uL capacity)
Gels
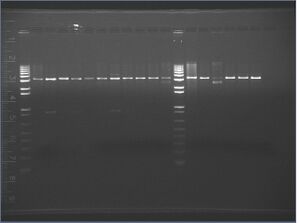
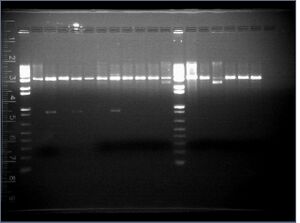

Analysis
It seems like we got KaiA but not KaiB, from the digests above. When the plasmids are cut with only one of the two restriction enzymes (due to a backwards insert in the vector), there should be a band about 3kb; that is seen in every lane. Interestingly, KaiA (which is 855 bp) shows up on the gel as an 850 bp band, but the vector containing KaiA should have been 2 kb instead of 3 kb.
Because we feel that we should analyze this further, we will wait until the morning to figure out what to do.