IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-8-14
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
To-do
- Midiprep
Midiprep pSB4A3, J04500 (online instructions here, page 16)
- Insert:
Digest miniprepped KaiA/B/C in GeneArt vector with X-P- Gel-purify
- (maybe) Inoculate more KaiA/B/C in GA vector
- Backbone:
Digest J04500\S-P, \X-P (for Stage I constructs and BB high-copy)CIP treatment of the above- Gel-purify the above
Find and transform B0034
- KaiABC ligation:
- KaiABC\X-P + J04500\S-P (stage I)
- KaiABC\X-P + pSB1AK3 (well-formed BB) (we can purify this backbone from the J04500\X-P digest)
- KaiABC\X-P + B0034\S-P (stage I) (must wait until we grow up and miniprep B0034)
- Sequencing:
- Sequence miniprepped KaiA/B/C + GA vectors, natural KaiA
- Western blots:
- Ligate GFP dev\X-P and pSB4A3\S-P, transform (Wait until we've midiprepped more pSB4A3 and do another digest, because the pSB4A3\S-P from our digest assay was a curious size)
Inoculate GFP dev + pSB1A2
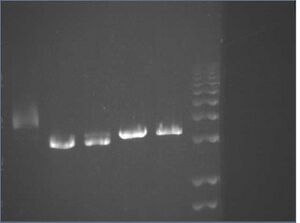
E-gel images of colony PCR of J04500\S-P + KaiA/B/C\X-P
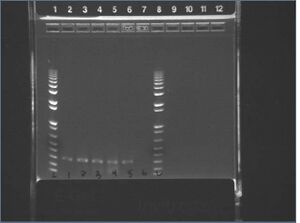
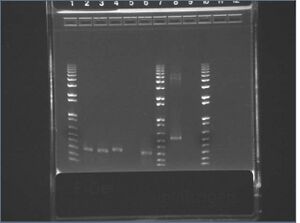
It appears that our vector self-ligated.
Digestion Assay for 8/14
| Label | DNA | Amount | Water | Enzymes | Amount | NEB buffer | BSA (100x) | Time | Master | Total | |
| Digests | 1 | J04500 | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 2 hrs | 1 | 25 µL |
| 2 | J04500 | 19.5 µL | 1.75 µL | SpeI & PstI | 0.5 µL, 0.5 µL | Buffer 2 (2.5 µL) | 0.25 µL | 2 hrs | 2 | 25 µL | |
| 3 | J04500 | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 4 hrs | 1 | 25 µL | |
| 4 | J04500 | 19.5 µL | 1.75 µL | SpeI & PstI | 0.5 µL, 0.5 µL | Buffer 2 (2.5 µL) | 0.25 µL | 4 hrs | 2 | 25 µL | |
| 5 | KaiA | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 2 hrs | 1 | 25 µL | |
| 6 | KaiB | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 2 hrs | 1 | 25 µL | |
| 7 | KaiC | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 2 hrs | 1 | 25 µL | |
| 8 | KaiA | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 4 hrs | 1 | 25 µL | |
| 9 | KaiB | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 4 hrs | 1 | 25 µL | |
| 10 | KaiC | 19.5 µL | 1.75 µL | XbaI & PstI | 0.5 µL, 0.5 µL | Buffer 3 (2.5 µL) | 0.25 µL | 4 hrs | 1 | 25 µL | |
| Master Mixes | 1 | - | - | 15.75 µL | XbaI & PstI | 4.5 µL, 4.5 µL | Buffer 3 (22.5 µL) | 2.25 µL | - | - | 49.5 µL |
| 2 | - | - | 7 µL | SpeI & PstI | 2 µL, 2 µL | Buffer 2 (10 µL) | 1 µL | - | - | 22 µL |
Add 5.5 µL of the corresponding master to each tube.
Transformation of B0034
We transformed B0034 (the Elowitz RBS) for use in one of our constructs. We plan to ligate it with KaiC for several of our Stage I experiments.
B0034 was rehydrated in 15 uL H2O and placed in the "Rehydrated Biobricks" container of the -20C freezer in the gel imaging room.
The part is on a pSB1A2 vector.
Transformations:
- 30 uL Top10 competent cells, 1 uL B0034
- 20 uL Top10 competent cells, 1 uL H2O (negative control)
Plates:
- B0034 in pSB1A2 in Top10 #1 (20 uL of SOC medium plated)
- B0034 in pSB1A2 in Top10 #2 (rest of SOC medium plated, ~260 uL)
- H2O in Top10 (50 uL plated) (negative control)
Inoculation of GFP device
We inoculated 5 mL of LB-Amp with GFP device from frozen stock. This will be used in preparation for tomorrow's Western blot tutorial.
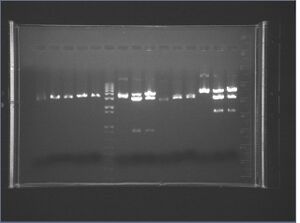
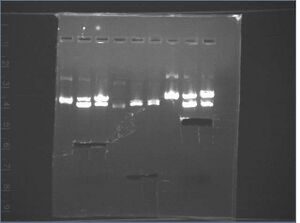
Gel image of J04500\S-P, J04500-X-P, KaiA/B/C\X-P
It appears the KaiB digest failed, and we also don't see the 220 bp insert from J04500\X-P. There's also a lot of undigested plasmid clearly visible in the Kai digests. I think we need to digest for longer and use a PCR machine instead of the water bath.
Midiprep
Notes:
- Reagents were added to the midiprep initially under maxiprep conditions (more amounts of buffers).
- The cylindrical containers were turned upside down to let the ethanol drain out.
- The ethanol was drained for 20 minutes.
- The final elution of DNA did not pick up an efficient amount of DNA, and therefore we performed three elutions of the DNA in 250 uL of water.
- Nanodropped amounts of DNA:
- J04500
- 97.6 ng/ul
- 34.1 ng/ul
- 31.5 ng/ul
- pSB4A3
- 36.4 ng/ul
- 33.3 ng/ul
- 6.1 ng/ul
- J04500