IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-7-6
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Newer entries at the bottom.
Plasmid requests
We emailed Professor Golden and requested the following plasmids and their sequences:
- pAM2195
- Chloramphenicol resistance [NS 2.1]
- PpsbAI::Lux AB [NS 2.1]
- PpsbAI::Lux CDE [NS 2.1]
- Ampicillin resistance [for selecting in E. coli, not on a neutral site]
- pAM1579
- Kanamycin resistance [NS 2]
- pAM2314
- Spectinomycin resistance [NS 1]
- Streptinomycin resistance [for selecting in E. coli, not on a neutral site]
Game plans
Preface
Recall that we want to run two experiments in cyanobacteria:
- Control: A control experiment in which we insert the Lux AB/CDE genes under the psbaAI promoter (which is known to oscillate on a circadian rhythm), to callibrate our equipment and verify that the cyanobacteria's luminescence will oscillate. This will duplicate previous research.
- BB test: An experiment in which we knockout the wild-type KaiABC genes and insert our own BioBrick'd KaiABC genes, along with Lux AB/CDE from the control. This will verify that our KaiABC construct works.
Yesterday we came up with two plans for running these experiments.
Plan 1
Plan 1
PCC 7942 (unmodified)
/ \
/ \
Control: + AM2195 [NS 2.1] BB Test: + (AM2195 + BB'd KaiABC) [NS 2.1]
- Plan 1 workload
- Transformations
- PCC 7942 + AM2195
- PCC 7942 + modified AM2195
- (2) with wild-type KaiABC knockout
- Mutations
- Mutate AM2195 for BB compatibility
- Ligations:
- Ligate mutated AM2195 with BB'd KaiABC insert
- Transformations
Plan 2
Plan 2
PCC 7942 (unmodified)
|
|
Control: + AM2195 [NS 2.1]
|
|
BB Test: + (AM2314 + BB'd KaiABC) [NS 1]
- Plan 2 workload
- Transformations
- PCC 7942 + AM2195
- (1) + modified AM2314
- (2) + wild KaiABC knockout
- Mutations
- Mutate AM2314 for BB compatibility
- Ligations
- Ligate mutated AM2314 with BB'd KaiABC insert)
- Transformations
Comparison
Plan 1 has the advantage of running both experiments in parallel. However, we don't yet know whether neutral site 2.1 is large enough to accomodate the resistance cassete, LuxABC, and KaiABC (> 8kb size).
Plan 2 is conceptually cleaner, since we could reuse our control strain and keep separate functions on separate neutral sites. We also know that our inserts will fit. However, Plan 2 requires the control experiment to finish before starting the test (it's serial, not parallel).
Nick's suggestion
Nick suggested merging transformations 2 and 3 of each of the above plans by overwriting the wild-type KaiABC genes with our own BB'd KaiABC (plus resistance) via H.R., instead of knocking out the wild-type and inserting our genes at a neutral site in separate steps.
Nick's suggestion works better for plan 2 than for plan 1, since in plan 1 we would need to overwrite the wild-type KaiABC with our BB'd KaiABC plus Lux AB/CDE, which puts the Lux genes at a different position than the control, making the control less meaningful.
For plan 2, in contrast, we would be building on the control strain, so we wouldn't have to re-insert Lux at a different place.
Modeling
We want to make a model to analyze how the phosphorylation of KaiC changes as you add more KaiC.
The following are Nick’s ideas for the model:
D[p]/dt = α(t) – α[P] [P](t) = [P](t – delta t) + α(t – delta t) – α[P](t – delta t) α(t – delta t) = sin(t) α(t) = k
A T: add α (t – delta t) new objects to A Remove (at random B length (A) objects from A Update phosphorylated state of each member of A Calculate average phosphorylated states of A [P] = [0,0,0]
Hydration of delivered primers
Peng's 10 primers for extracting and site mutagenesis of KaiABC came in; they were each rehydrated into 250uL of dH20 for a stock solution. Then, 20mM of each was made for use.
In the 250uL stock the concentration is:
7942_KABC_extF: 94.08 nmol/mL 7942_KABC_extR: 37.96 nmol/mL 7942_KABC_crossF: 133.88 nmol/mL 7942_KABC_crossR: 192.64 nmol/mL 7942_KABC_pst1R: 154.16 nmol/mL 7942_KABC_pst1F: 150.28 nmol/mL 7942_KABC_pst2R: 153.64 nmol/mL 7942_KABC_pst2F: 168.2 nmol/mL 7942_KABC_eco1R: 145.84 nmol/mL 7942_KABC_eco1F: 138.56 nmol/mL
To make a 20mM concentration with ~50 µL volume for each, add:
7942_KABC_extF: 8uL stock, 37.63uL h20 7942_KABC_extR: 16, 30.37 7942_KABC_crossF: 6, 40.16 7942_KABC_crossR: 4, 38.6 7942_KABC_pst1R: 6, 46.28 7942_KABC_pst1F: 6, 45.084 7942_KABC_pst2R: 6, 46 7942_KABC_pst2F: 6, 50.46 7942_KABC_eco1R: 6, 43.75 7942_KABC_eco1F: 6, 41.568
PCR from dead PCC7942
The goal here is to extract the kaiABC gene cluster from our dried out PCC7942 plates. Result: fail =(
Each person did 3 samples; 2 experimental and one control.
The experimental had:
5uL template 5uL 10x Buffer 1uL dNTP (10mM ea) 1uL Primer (extF) 1uL Primer (extR) 0.5uL HotStarTaq 36.5uL dH20
The run cycle was:
#94C, 15' #94C, 30" #56C, 30" #72C, 1.5' #Cycle to step 2, 7x #94C, 30" #65C, 30" #72C, 1.5' #Cycle to step 6, 30x #72C, 5' #4C hold
Egel results show nothing good...
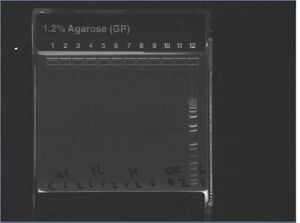
Incubator status
- Added
- PCC 7942 streak #1 plate
- PCC 7942 streak #2 plate