IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-7-18
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
DNA Miniprep of topo+kaiabc clones
With the Topo prepared clones in e.coli Hetmann created:
1: kaiabc+topo #1 2: kaiabc+topo #1 3: kaiabc+topo #1 4: kaiabc+topo #1 5: kaiabc+topo #2 6: kaiabc+topo #2
I aliquoted 4mL of each topo clone into its seperate vial and conducted a DNA miniprep based on the Qiagen miniprep kit 6/05 handbook p22-23. Used old Qiagen miniprep kit (had very little P2 buffer in it); new one did not have RNAase added. #1,4,and 5 did not have as many cells as 2,3, and 6. Used centrifuge for the large centrifuge tubes; 4.4k for 10 min.
note: tubes 4,5,6 may have been a bit contaminated by either 1,2, or 3; during loading of first centrifuge, but most likely okay.
The final reactions are stored in blue 1.5 tubes marked (number, kaiabc topo DNA). used 30 uL extraction.
DNA nanodrop data:
1: 153.3 ng/uL 2: 14.0 ng/uL 3: 14.4 ng/uL 4: 8.9 ng/uL 5: 12.5 ng/uL 6: 27.2 ng/uL
Weirdly enough, sample 1 and the other ones fell within 260/280 as 1.8-2.0... did NOT dilute sample 1.
Storage of topo+kaiabc clones
For all 6 topo clones, I aliquoted 666uL of each into a cryogenic vial, and added 666uL of 50% glycerol. Each is in a vial at -80C, in a cap with blue writing and dated.
Digest of the topo+kaiabc clones and the one we were using the past week
In order to do a pre-sequence test to see if our template is correct, I did a restriction digest using the pstI enzyme. Based on this Media:Pcr2topoblunt.pdf file, if we cut at the pst1 site only we can do three diagnoses. The following is what I look to test.
- If there is no kaiABC in the vector, we will get a 3519 bp fragment.
- If there is no kaiABC in the vector and kaiABC freeflowing, we will get a 500 bp fragment, a 2390bp fragment, and a 4519 bp fragment
- If there is kaiABC in the vector, we will get a 2390bp fragment and a 4138bp fragment
- If there is random crap in the vector, the bands will be different; a 3kb band prehaps?
- Positive control: reactions itself?
- Negative control: no DNA
Granted, using SpeI or double digesting may provide more info, but I think we're out of SpeI and ... yeah.
Per reactions 2,3,4,5,6 the amount in a 50uL rxn (gives ~140-270ng of DNA/rxn depending on concentration):
10uL DNA 5uL 10x Buffer3 1uL PstI 0.5uL 100x BSA 33.5uL dH20
For reaction 1 the amount in a 50uL rxn (gives 300ng of DNA):
2uL DNA 5uL 10x Buffer3 1uL PstI 0.5uL 100x BSA 41.5uL dH20
For reaction "Tp" the amount in a 50uL rxn (gives 300ng of DNA, was template we were using last week):
5uL DNA 5uL 10x Buffer3 1uL PstI 0.5uL 100x BSA 38.5uL dH20
For negative:
5uL 10x Buffer3 1uL PstI 0.5uL 100x BSA 43.5uL dH20
Master Mix was preformed for all samples: combining BSA, Buffer3, PstI, and dH20.
The run time is:
37C for 6 hours 80C for 20 minutes 4C for forever
The run is being done on the PCR machine #1. Will be done by 1030AM 7/18.
Repeat of colony PCR (see 7/10)
Assuming that our digests are not turning out good, I repeated the colony PCR step from cyanobacteria. Biggest change here is the use of solid monocolonal colony intstead of liquid polyclonal. Lysing by thermocycler @ 95C @ 5min.
The protocol is:
1 experimental, 1 negative (no template), and 1 BB test.
The experimental/negative had:
5uL/0uL colony DNA 1.5uL 20mM dNTP (new) 1.0 uL extR 1.0 uL extF 0.5uL HotStarTaq (N) 5uL 10x buffer 41.5/46.5 dH20
The BioBricks test, which looks for contamination in the primers, had:
1.5uL dNTP 1.0 uL RF 1.0 uL (reverse) 1.0 uL HotStarTaq (N) 5 uL 10x buffer 41.5uL dH20
They are in 0.2mL miniPCR tubes in PCR#6. (3 samples total) Tops in blue.
Repeat of 7/17 transformation
I (Jeff) plated yesterday's transformants on the wrong plates, so I redid the Topo ligation and transformation. I used 30 µL, 15 µL, and 15 µL of competent cells for the experimental, negative, and positive controls.
I plated the transformants on 6 plates (1 negative control, 1 positive control, 4 experimental) at around 1 PM.
Gels
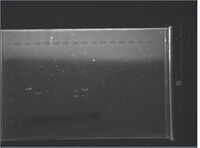 Click for legend |
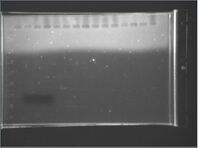 Click for legend |
 Click for legend |
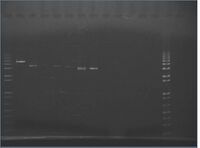
Primers
We received primers today for sequencing our KaiABC extract and extracting KaiA, KaiB, and KaiC separately. We hydrated the primers and made dilutions as recorded below:
Sequencing primers
| Primer | Stock concentration | Stock volume | Dilution concentration | Dilution volumes |
| seqA1 | 179.64 mM | 250 µL | 20 mM | 5.57 µL stock + 44.43 µL H20 |
| seqA2 | 189 mM | 250 µL | 20 mM | 5.29 µL stock + 44.71 µL H20 |
| seqA3 | 205.64 mM | 250 µL | 20 mM | 4.86 µL stock + 45.14 µL H20 |
| seqA4 | 155.2 mM | 250 µL | 20 mM | 6.44 µL stock + 43.56 µL H20 |
| seqA5 | 190.8 mM | 250 µL | 20 mM | 5.24 µL stock + 44.76 µL H20 |
| seqA1_RC | 201.72 mM | 250 µL | 20 mM | 4.96 µL stock + 45.04 µL H20 |
| seqA2_RC | 172.88 mM | 250 µL | 20 mM | 5.78 µL stock + 44.22 µL H20 |
| seqA3_RC | 192.88 mM | 250 µL | 20 mM | 5.18 µL stock + 44.82 µL H20 |
| seqA4_RC | 218.12 mM | 250 µL | 20 mM | 4.58 µL stock + 45.42 µL H20 |
| seqA5_RC | 199.4 mM | 250 µL | 20 mM | 5.01 µL stock + 44.98 µL H20 |
Extraction primers
| Primer | Stock concentration | Stock volume | Dilution concentration | Dilution volumes |
| exAF | 66.52 mM | 250 µL | 20 mM | 15.03 µL stock + 34.97 µL H20 |
| exAR | 100 mM | 328.2 µL | 20 mM | 10 µL stock + 40 µL H20 |
| exBCF | 52.52 mM | 250 µL | 20 mM | 19.04 µL stock + 30.96 µL H20 |
| exBCR | 100 mM | 309.4 µL | 20 mM | 10 µL stock + 40 µL H20 |
| exBOR | 100 mM | 165.7 µL | 20 mM | 10 µL stock + 40 µL H20 |
| exCOF | 68.36 mM | 250 µL | 20 mM | 14.63 µL stock + 44.43 µL H20 |
Reinoculation of the Hetmann Six
We reinoculated Hetmann's 6 plated cultures in 5 mL of LB (w/ 2 µL Kan == 20 µg/mL), using 10 µL liquid culture from the frozen stocks that Peng made in the early morning.