IGEM:Harvard/2006/Cyanobacteria/Notebook/2006-7-14
<html><style type='text/css'> .tabs {
font-size:80%;
font-weight:none;
width: 100%;
color: #FFFFFF;
background:#FFFFFF url("/images/5/54/DarkgreenTab-bg.gif") repeat-x bottom;
}
.tabs li {
background:url("/images/3/36/DarkgeenTab-left.gif") no-repeat left top;
}
.tabs a,.tabs strong {
background:url("/images/d/d3/DarkgreenTab-right.gif") no-repeat right top;
color:#FFFFFF;
padding: 3px 10px 3px 4px;
}
.tabs strong{
color:#CCFF00;
background-image:url("/images/b/b1/DarkgreenTab-right_on.gif");
}
.tabs a:hover{
color:#66FF00;
}
</style></html>
Colony PCR from 7/13
Failed... negative had stuff too. The result is from a 1.2% e.gel run for 15min.
We were supposed to get the following sized products:
- 3-9: 209bp
- 10-7: 370bp
- 8-5: 2402bp
- 6-4: 80bp
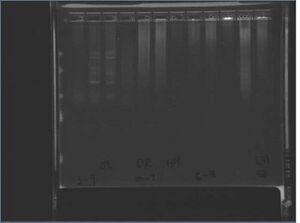
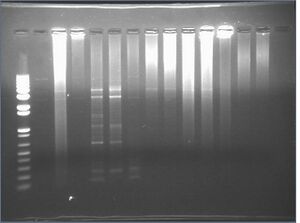
2: DR 8-5
3: HH 8-5
4: JL 8-5
5: DR 3-9
6: HH 3-9
7: JL 3-9
8: DR 6-4
9: HH 6-4
10:DR 10-7
11:HH 10-7
12:HH neg
13:JL neg 3-9
14:DR neg 6-4
15:empty
Although it looks like we got bands, all negative had junk in it; did a master mix, so most likely something went wrong there.
Another PCR attempt
We decided to try another PCR using the Topo + KaiABC plasmid Peng miniprepped yesterday. This time, each of us would make his own master mix. Since we accidentally left our Vent polymerase out on the bench yesterday, we decided to split and do half our reactions using Vent and half using Hotstar. We also decided to dilute the DNA template since our last gel had smearing. Our template is now 500x diluted instead of 100x. This should give us somewhere around 6 ng absolute mass of DNA, if the nanodrop is accurate.
Each person made 5 reactions: 4 using each pair of mutations primers, and 1 negative control with H20 instead of template.
Peng and Jeff used Hotstar Taq, while Hetmann and David used Vent.
Reaction mixture
Each reaction contained:
| PCR mutagenesis | |
| 0.1 μL template or H20 | |
| 5 μL 10x buffer | |
| 1 μL 10 mM dNTP | |
| 1 μL Primer 1 | |
| 1 μL Primer 2 | |
| 0.5 μL Taq (Vent or Hotstar) | |
| 41.4 μL dH20 | |
Protocol for mixing
- To each tube, add:
- 5 µL H20
- 1 µL primer 1
- 1 µL primer 2
- Make the following master mix:
- 9 µL H20
- 1 µL template DNA
- Add 1 µL of the above master to each experimental tube, and add 1 µL of H20 to the control (there should be leftover master mix).
- Make the following master mix:
- 30 µL buffer
- 6 µL dNTP
- 213 µL H20
- 3 µL Taq
- Add 42 µL of the above master to each tube