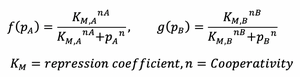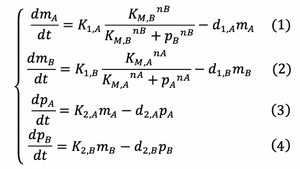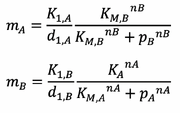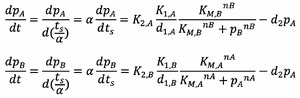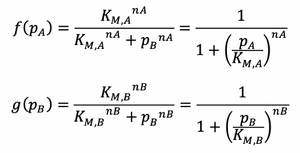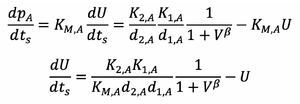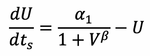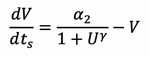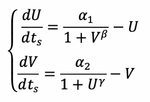ICSynBio:TSModelDerivation
Toggle Switch Model Derivation
The Model used by Gardner et Al to describe the genetic construct of the Toggle Switch can be derived from first principles.
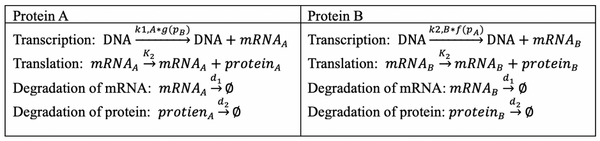
Chemical Reactions
We start with the Chemical equations for Gene Expression (Central Dogma of Biology):
Some Assumptions we make are:
- Transcription Rate can be given by following hill functions:
- The Translation Rate parameter is constant.
- The mRNA and Protein degradation parameters are constant.
ODEs of the system
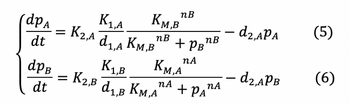
The Law of Mass Action allows us to yield the ODE’s below that describe the rate of change of protein and mRNA concentration:
The definitions of the variables and parameters in above ODE's are:
Quasi-Stationary Approximation
We can also make the Quasi-Stationary approximation for mRNA concentration. This is because we know that the concentrations in our system will go to a steady state, and that the mRNA concentrations reach a steady state faster compared to the Protein concentrations.
Rearranging Equations 1 and 2 Yields:
Substituting this result into Equations 3 and 4 gives:
Normalisation of the system
Our aim is to get a more parsimonious model that can be described by fewer parameters. Further normalisations` and simplifications can be made to this.
We know that protein degradation rate is the same for both protein A and protein B. This is because Proteins are stable and their degradation rate is equal to the dilution rate.
We first define a new time scale:
Substituting this into equations 5 and 6 gives:
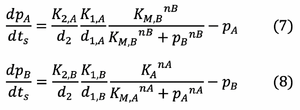
And rearranging this gives:
The following rearrangement can be made for the hill function expressions:
We can define the following dimensionless concentrations:
As well as this to get the same equations as Gardner et Al we use ![]() and
and ![]()
Therefore equation 7 can be written as:
We then define a lumped parameter ![]()
Substituting this into the above equation yields:
Similarly for equation 8 and defining ![]() gives:
gives:
The system
Therefore the same Equations that Gardner et Al used have been derived from first principles:
Interpretation of the Variables and Parameters:
- U and V are the dimensionless concentrations of the two proteins that mutually repressing each other, normalised by the Km (the repression coefficient), of the respective proteins.
- The dimensionless timescale has been normalised by d2, the degradation rate of the proteins.
- alpha1 and alpha2 are lumped parameters that define the maximum steady state concentrations of the proteins normalised by the Km of the respective proteins.
- Beta and Gamma are the hill function coefficients, which represent the cooperativity- the binding of chemicals to the promotors.
Further Links
Further analysis of this system can be found here