Endy:Dedicated systems/Virtual machines
Back to Dedicated systems
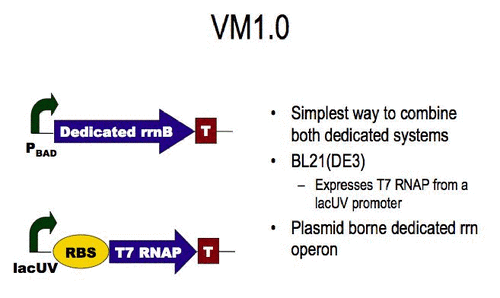
VM1.0
Introduction
We are working to build and test biological virtual machines. A virtual machine is a standard interface between a platform (such as a specific computer architechture) and an arbitrary system (such as a software application). A biological virtual machine allows any engineered biological system that conforms to a particular specification to function in any cellular chassis for which a virtual machine exists.
A simple implementation of a biological virtual machine is shown on the right. Initial testing suggests that this virtual machine is a functional way to produce a specific protein, see the results section below. A plasmid encoding dedicated ribosomes were obtained from Chris Hayes based on the earlier Brink paper. T7 RNA polymerase is used as a dedicated transcription system.
BL21(DE3) already contains a dedicated transcription system. Together with pCH1497-ASD1, this cellular chassis becomes BL21(VM1.0). Any engineered system whose protein synthesis is controlled by genetic elements conforming to the VM1.0 specification will function in this chassis.
BL21(DE3) was obtained from Sri Kosuri. If was transformed with pCH1497-ASD1 that was obtained from Kathleen McGinness who obtained it from Chris Hayes. The resulting strain is termed BL21(VM1.0) and can be obtained from Barry Canton. For location information, see here.
Reporter Device
- Contains T7 polymerase specific promoter and terminator
- Contains an RBS that is only recognized by ribosomes with a specific mutation in the 16s rRNA.
- Currently on pSB1A2.
Measurement Protocol
Overnight Culture
Slightly modified from Bacterial cell culture.
- Streak a fresh plate. Either because of the tetracycline or because of the leaky expression of the dedicated ribosomes, colonies seem to die on the plates quite fast. For best results use a plate that is less than a week old.
- Pick single colonies into 2ml of media into a 17mm culture tube. Normally Neidhardt EZ media is used for growing BL21(VM1.0) but supplemented M9 media also works.
- Culture at 37o in a roller for approximately 12hours. Early results suggested that subsequent growth of the cultures was more likely to proceed if BL21(VM1.0) was not allowed to grow into stationary phase. However it may be that the only important parameter is how long the cultures are left to grow before induction of the dedicated machinery. This point needs to be confirmed. Regardless, it is probably better not to let the cells reach stationary phase to minimize changes to their physiology over the course of the experiment.
Measurement growth phase
- Dilute into fresh media with a dilution of 1/500. 1/250 or 1/1000 dilutions are also suitable, a dilution of 1/500 simply means that the cultures will be ready to be induced within 3-4 hours.
- Once the cultures reach on OD of ~0.1, add inducers as appropriate. Again, the choice of 0.1 is somewhat arbitrary although it is necessary to allow cell density to reach a detectable level prior to induction as cell growth stops after ~3hours of induction.
- Fluorescence should be evident within minutes of induction.
Results
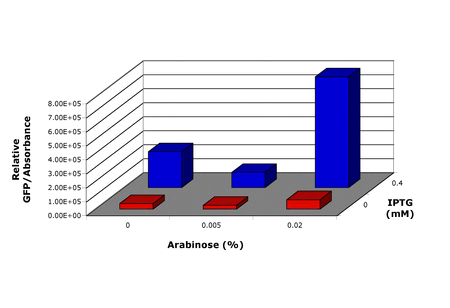
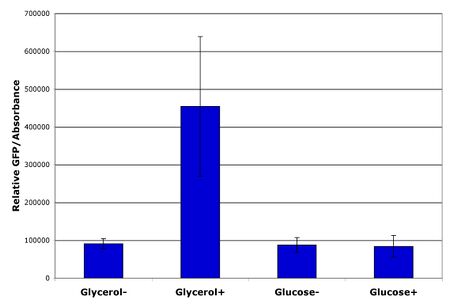
Figure 1 shows the results from the first experiment to test GFP production using VM1.0. IPTG was used to induce the dedicated transcription system and arabinose was used to induce dedicated translation. The results showed qualitatively that the dedicated systems were working in combination. However, these results were hard to repeat. Cultures did not grow, or did grow but did not produce fluorescence when induced, and plates became old rapidly.
More recently, consistent results have been obtained obtained. Performing the first dilution at a lower OD (~0.6) seemed helpful and allowing the cells approximately 5 subsequent hours before inducing the dedicated systems produced the results shown in Figure 2. The figure shows the GFP level as measured by a plate reader normalized by the absorbance of the culture. VM1.0 containing the reporter construct BBa_E7103 was grown in the presence of 0.4% glycerol or glucose and either with or without inducers of the dedicated systems. Replicates from three different colonies were grown for each condition. The error bars represent +/- one standard deviation of the replicates. The results show that glucose effectively shuts off the dedicated systems as is expected since they are controled by catabolite repressible promoters.
Now that a reliable protocol for using the virtual machine exists, we are currently performing more quantitative analysis of the behavior of VM1.0.
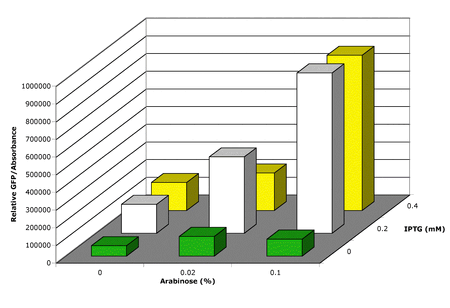
- Latest results, shown in Figure 3, suggest that 2mM IPTG is sufficient for full induction of dedicated transcription.
- In addition the results suggest that using 0.1% Arabinose leads to induction of a larger fraction of the population and less variability in the population fluorescence level.
- There appears to be more leakiness or non-specific action in the dedicated translation system than the dedicated transcription.
It would appear that if dedicated ribosomes are induced a number of hours prior to inducing dedicated transcription that GFP accumulates faster than when both sets of dedicated systems are induced at the same time. This is what might be expected. However, in the long term, net GFP accumulation is greater when the induction of both systems occurs simultaneously. Furthermore, the prior induction of dedicated translation does not affect the accumulation rate of GFP when the reporter device uses the cellular chassis' ribosomes. Net GFP accumulation is again higher when both sets of systems are induced simultaneously. It would appear that having dedicated ribosomes in the cell does have a long term effect on the supplies available to the system even though the two different induction conditions do not affect final cell density.