Endy:Chassis engineering/Orthogonal protein synthesis
Motivation
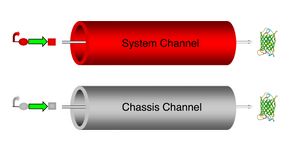
We are working to reduce the coupling between engineered biological systems and their cellular chassis. This coupling leads to unexpected system behavior and perturbed cellular function. A major source of the coupling between system and chassis is that both share the same transcription and translation processes. We are working to implement an orthogonal protein synthesis channel that is solely used by an engineered biological system and does not perturb cellular function.
If this orthogonal channel can be standardized and designed to function in multiple cell types, then it forms the basis of a biological virtual machine(1.0, 2.0).
Currently, we are working to enable dedicated transcription and dedicated translation in an E. coli strain MG1655 chassis. Details on this work can be found on the following pages -
Demonstrating the usefulness of dedicated systems
- Measure specificity of combined dedicated systems
- Demonstrate decoupled function of the system and the cellular chassis
- GFP accumulation as chassis make the transition from log to stationary phase should indicate whether system performance is less affected by the change in the chassis state.
- Suddenly turning on the high level protein expression of a system should reduce growth rate of the chassis when the system uses dedicated systems than when it does not. Initial experiments have suggested that this might only be true in a minimal media such as M9.
First generation reporter devices for the dedicated transcription/translation systems
| General Translation | VM Translation | |
|---|---|---|
| General Transcription | I7101 |
I7102
|
| VM Transcription | E7104 |
E7103 |