Biomod/2015/StJohns:Approaches
http://openwetware.org/images/f/f9/Lukemanlab-toehold-conga-nanny-header.jpg
Approaches
Claw Synthesis
Vanilla Claw was equipped with sticky ended regions; we planned on demonstrating binding via gel shift and TEM imaging. It was decided that aside from those two techniques it would also be helpful to illustrate virus-capsid binding and conformational change through an energy transfer technique (FRET). For that, NClaw was designed differing only from "Vanilla" claw by the presence of the acceptor and donor modified staples and their adjacent staples. As mentioned in the previous section: one acceptor site and two donor sites near the hinge region of the claw. n the initial approach, claw was designed to have one virus-binding site on each arm. After binding to capsid was not successful (see Capsid section below), we decided to add multiple binding sites to each arm of the arm, varying from 9 (3/arm) to 15 (5/arm). It was seen that the 5S Claw showed more binding (See Figure 14 in Results). In this case, more binding was shown by polymerization on the gel. Though binding efficiency was increased, there was now an even greater problem with claw aggregation and polymerization.
In an attempt to limit polymerization, the TClaw was assembled, equipped with tether staples extending from one arm to the next, effectively rigidifying the claw structure. 22nm, 31nm, 40nm, and 49nm tether lengths were originally designed (10 base pairs= 3.5 nanometers). 22nm length was the minimum calculated distance needed between each arm for the capsid to fit within. At this point, due to tether location, one of the the two FRET acceptor sites was replaced by tether attachment.. Initially the same assembly protocol was used in order to attach the tethers. This consisted of combining all strands in buffered solution, denaturing the strands by raising temperature to 90C. Afterwards, allowing them to anneal back to room temperature. This resulted in polymer formation, prior to capsid binding. (Figure 15 in Results) Afterwards we decided to try various anneal protocols; of which capsid anneal protocol (See Procedure) decreased the polymerization. This required raising the temperature to only 37C, and allowing annealing to room temperature. (Figure 16 in Results)
At this point binding of tethers was still not clearly seen, since the tethers could just have stayed in solution, not having bound to anything. Using fluorescent HEX strand, with regions complementary to those on the single stranded tether strands (31nm, 40nm, and 49nm), would illustrate tethers binding to claw, without the use of SYBR stain. By adding excess tether and HEX strand it was seen that compared to control group of only Tether and HEX, that when bound the amount of starting material decreased; possibly indicating binding. (See Figure 17 and 18 in Results section, Post SYBR stain and pre SYBR respectively). The CY3 Fluorescent Strand was then tried, due to its lower limit of detection. Figure 19 and 20 in Results (respectively post SYBR and pre-SYBR) illustrate that the tether strands were attaching to the claw.
Capsid Binding
The capsid/origami binding protocol was adapted from the protocol used by the Stephanopoulos group that worked on binding with triangle D.O., annealing from 37°C to 4°C at 1°C/min.[3]). However, the results of virus capsid binding experiments with claw were less than favorable; no significant band shifts occurred that would signify a binding the X MDa capsid to the claw. Many experimental conditions were tested to obtain binding, varying a series of anneal conditions as well as gel conditions. They include capsid/origami stoichiometry, Mg2+ concentrations of the solution during anneal, gel run temperature, and the sequence of the ssDNA used at the binding sites of the claw. Figure 11 is a summary of what was done in varying conditions for these tests. Figure 12 shows a gel image representative of all the images obtained from the various tests from Fig. 11; smeared lanes for samples of capsid+claw were common for these gels. The tests in Fig. 11 used DNA-modified MS2 capsids that were at least a year old from receive date, so our collaborators from Berkeley sent another batch of modified capsids, which were used in our most recent binding experiments.
Plagued by unsuccessful binding with our original claw design (Figure 12 in Results), we decided to take a step back and try binding with triangle D.O., the design used in the original virus-capsid binding protocol. Triangle origami was made with 5 binding sites on each side, with binding experiments immediately following. With this design, a noticeable band shift occurred, signifying capsid bound to origami (Figure 13). Hoping to mimic this success using the claw design, we looked to see what was different, other than the main difference in origami design. The only difference in the two systems was the number of binding sites on each arm/side. With this experiment, the number of binding sites per arm became another variable to test.
Even with multiple binding sites per arm on the claw, we could not get a band shift as clean as the experiment using triangle origami (Figure 13). We concluded that complex polymers form during the annealing process, resulting in bands smearing upwards in our gel images. One of the solutions we came up with to limit polymerization of capsids and origami was to restrict the mobility of the claw. To do this, we modified the design to allow ssDNA tethers to be added, which would connect adjacent sides of the arms of the claw.
The current goal of the project is successfully optimizing the conditions for capsid binding with the tethered claw design. An approach similar to the initial one using the original claw design will be taken to optimize binding. The conditions that will be varied include capsid/claw stoichiometry, Mg2+ concentration, anneal time, anneal concentration, and the like.
FAB’ Work
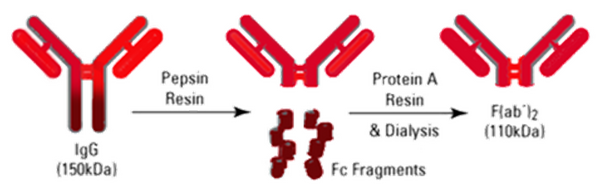

Our approach to improve binding was to modify the DNA claw with specific binding sites for the wild-type MS2 virus capsids. To make Anti-MS2 Fab’, we have shown IgG1 or IgG2a can be digested. Comparing Figure 20 and Figure 24, In Figure XX Gel it is clear that IgG2a has fewer digest by-products. For IgG2a, we use pepsin, a nonspecific endopeptidase, to digest the Fc portion of IgG2a to yield F(ab')2. This fragment is composed of a pair of Fab’ units connected by two disulfide bonds.
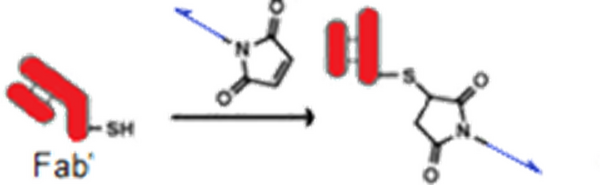
After making the F(ab’)2 we reduce the disulfide bond in the hinge region of the antibody making Fab’ (~50 kDa).
Amine-modified DNA has been coupled to an anti-MS2 Fab’ via a maleimide linker. (DNA oligo in blue not to scale)
Upon pursuing this project idea, the first reaction in the sequence; that of digesting the IgG antibody into F(ab’)2, was completed with ease using the Pierce F(ab’)2 Micro Preparation Kit purchased from Thermo Fisher Scientific. The second step was more challenging to go from F(ab’)2 to Fab’ required trials with differing concentration and temperatures. This kit utilizes the enzyme pepsin.
Work has been done to test Fab fragments rather than Fab’ fragments but wasn’t further pursued, as the isolated Fab yields were okay but when work to transaminate and then conjugate oxyamine or hydrazine PEGs and DNA resulted in vanishingly small yields. On account of this, we focused on Fab’ and maleimide chemistry rather than Fab.
2 other enzymes were also tested on 3 subtypes of IGG and either did not make Fab’ efficiently or overdigested the antibody to Fab fragments instead. Alternate antibodies – IgG1, IgG2b, and polyclonal IgG2a - were also tested to make these Fab’ fragments. IgG1 was used more than the others, but similarly to them had longer reaction times with no improvements to yield and were thus discontinued, but we have shown still yield the desired Fab’ (Figure 23). Three conditions were ran on a 12% SDS PAGE shown in Figure 20 and from then forth, condition 1 (50mM 2-Mercaptoethylamine•HCl @37°C). Following that moving into conjugation of the antibody fragment with a single strand of DNA meant testing both before continual attempts to make sure the elements themselves were functional. The smearing in Figure 21 and the smearing in Figure 22 was interpreted as functional and reactive DNA and antibody.
Following this, conjugation reactions were tried in many different conditions and factors shown in Figure 24. The end result was the band in Figure 25 that seems to be the conjugate, about ~10kDa above the 50kDa band for Fab’, which is plausible as the DNA used is about this weight.
Originally it was assumed that 2-5 equivalents of DNA to Fab’ should work but we have concluded from Figure 24 that 250:1 and the other listed conditions for test #5 were the best for this reaction.
After the extensive testing of reaction time, stoichiometry, concentration, and reagent batches always resulted a low yield; too low to make testable amounts of conjugate to attach to claw. Thus conjugating this antibody fragment to the claw will no longer be pursued as a prototype for sensing clinically relevant viruses.
We are currently pursuing aptamer recognition of virus surfaces. This aptamer will also be bound to the claw and tested for binding capabilities.
Techniques
- Atomic Force Microscopy (AFM)
- Gel Electrophoresis
- Transfer Electron Microscopy
- Fluorescent Resonance Electron Transfer
- SDS Page