Biomod/2012/Harvard/BioDesign/L-DNA layer
<html>
<head>
<link href='http://fonts.googleapis.com/css?family=Open+Sans' rel='stylesheet' type='text/css'>
</head>
<style>
/*BUT ACTUALLY*/ body {
font-family: 'Open Sans', sans-serif; overflow-y: scroll;
}
.container {
background-color: #ffffff; margin-top:0px
} .OWWNBcpCurrentDateFilled { display: none; }
h5 {
font-family: 'Open Sans', sans-serif; font-size: 11px; font-style: normal; text-align: center; margin:0px; padding:0px;
}
- column-content
{
width: 0px; float: left; margin: 0 0 0 0; padding: 0;
} .firstHeading {
display:none; width:0px;
}
- column-one
{
display:none; width:0px; background-color: #ffffff;
}
- globalWrapper
{
width: 900px; background-color: #ffffff; margin-left: auto; margin-right: auto
}
- content
{
margin: 0 0 0 0; align: center; padding: 12px 12px 12px 12px; width: 876px; background-color: #ffffff; border: 0;
}
- bodyContent
{
width: 850px; align: center; background-color: #fffffff;
}
- column-content
{
width: 900px; background-color: #ffffff;
}
- footer
{
position: center; width: 900px;
} @media screen {
body { background: #000000 0 0 no-repeat; /* changed default background */ }
}
- menu
{
position: fixed; float: left; width: 180px; padding: 10px; background-color: #FFFFFF;
}
- pagecontent
{
float: right; width: 620px; margin-left: 300px; min-height: 400px
}
- toc { display: none; }
/*Expanding list*/ ul { list-style: none; }
- exp li ul { display: none; }
- exp li:hover ul { display: block; }
- exp li a:active ul { display: block; }
a:link {color:#FF6060;} a:visited {color:#FF6060;} /* visited link */ a:hover {color:#B24343;} /* mouse over link */ a:active {color:#B24343; } /* selected link */
</style> </html>
L-DNA Layer Design
Connecting to the Template
When deciding how to connect the L-DNA strands to the template, we thought of a couple ways: either through a D-DNA handle, or through direct covalent linkage.
The handle method would be using a universal D-DNA handle, one strand attached to the tempalte and the complimentary strand attached to the L-DNA strand. This would make the L-DNA strand, partially L-DNA (for the SST layer) and partially D-DNA (the handle). But this would be okay as during detachment, which could be done through an enzyme, the D-DNA handle would get digested as well. One issue was that if the handle was attached to the end of the strand, the strands would need to be annealed - end to end connections may lead to complications if one end of the tile is already bound.
The second design, covalent linkage, would solve this problem. Using click chemistry, a reliable organic reaction that can covalently link two modified DNA strands, a connection could be made even from the middle of strand to another. This would free up all 4 ends for forming the SST layer. The downside is that this would require extra organic synthesis, adding complexity to the procedure.
Our team decided to use the handle method as this was a very clean design. We continued to explore click chemistry, even running a couple of experiments to test if we could - but did not continue to pursue that path after the first reaction.
L-DNA Sequence Design
We optimized two L-DNA SSTs with 8 total domains (shown below) so that they would assemble into orderly and infinite ribbons as shown. In the figure below, red and blue SSTs represent the 2 different tiles and complementary domains are designated as a and a'.
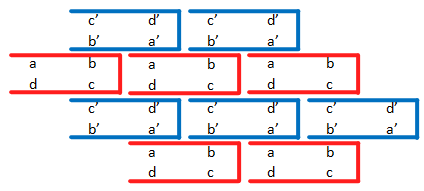
Sample tiling of 2 L-DNA SSTs (red and blue)
We did not need to optimize the L-DNA tiles to prevent interaction with the D-DNA canvas, since L-DNA and D-DNA cannot interact. However, it was necessary to optimize the strands for a lower melting temperature. This was an important step, since the L-DNA strands must bind to each other in a second annealing step, after the template canvas and handles have already annealed, without any of these structures dissociating. Thus, the melting temperature of the L-DNA strands with each other must be significantly lower than the melting temperature of the D-DNA template canvas and of the handle strand.
We created 6 initial pairs of strands with randomized sequences with limited GC content. We then analyzed each of these designs with NUPACK and Oligo Analyzer to check for low melting temperature and minimal interaction outside of the desired complementary helices.
| Design | Domain 1 | Domain 2 | Domain 3 | Domain 4 | Melting Temp. |
|---|---|---|---|---|---|
| 1 | AGACACAGA | AAGTAATGCA | ATACATCTTAA | AACAATTTAGAA | 33.7°C |
| 2 | ACTCGGATG | AACAGAATCA | AGAATGTAAAA | AGTATCAATAAG | 35°C |
| 3 | ACAATCAGA | CAAATAGGAA | CTAAAATGAAT | ACTTAGATAATA | 35.6°C |
| 4 | ATGATTTGA | AGTAAGTTAA | ATAGAAAAGTA | ATAGATAGTATA | 33.3°C |
| 5 | AAGAATGAT | AAGAAAAGTT | ATAGAAAGTTA | AGATATAAGATA | 34.1°C |
| 6 | CTCTAACAC | CATATCTTGT | AATACAATATC | TTATACTTTATC | 34.6°C |
D-DNA Analysis
We discarded design 2 for having too many intrastrand interactions and ordered the other 5 strands in D-DNA for testing.
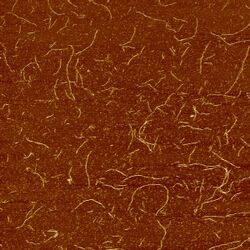
We annealed these sets of strands and imaged the results using AFM. The images below show structures formed by design 3. Rather than forming an infinite sheet, the strands fold into long tubes because of the natural curvature of the sheets.
Design 3 (both ribbon strands) imaged at 2μm (left) and 1μm (right)
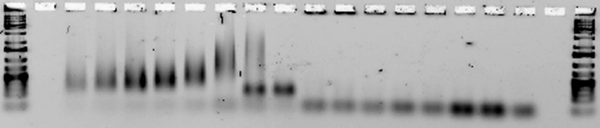
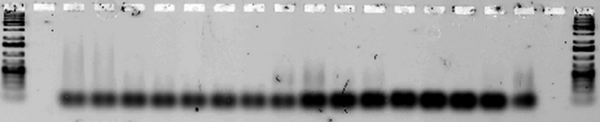
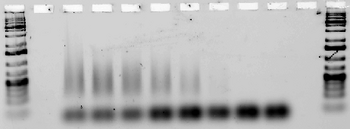
Next, we tested for annealing at a range of temperatures between 30 and 44.5°C to determine the melting temperature of each design(the lowest temperature at which the strands fail to anneal. (Note that in some lanes, e.g. for Ribbon 3, the strands aggregate so much that they remain in the well. Other lanes, e.g. those for Ribbon 5, contain a very faint smear.)
Ribbons 1 & 3, Temperature optimized
| ||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||
Ribbons 4 & 5, Temperature optimized
| ||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||
Ribbon 6, Temperature optimized
| ||||||||||||||||||||||||
| ||||||||||||||||||||||||
To limit cost, we chose to only order designs 3 and 5 in L-DNA. These designs both showed low melting temperature and the expected tube structure on AFM.
L-DNA Analysis
We ordered designs 3 and 5 in L-DNA for the final stages of testing. We did not test their hybridization with D-DNA as literature says D-DNA and L-DNA do not hybridize and we did not see any evidence of such.
| Small SST Canvas >> |