Biomod/2011/PSU/BlueGenes/overview
|
Home ::: Overview ::: Methods ::: Results ::: Application ::: Literature ::: Team |
OverviewProject GoalThe goal is to develop a computational algorithm for structural characterization of synthetic DNA. Successful completion of the goal would allow for fluctuation calculations of any synthetic DNA structure so long as it is in PDB (protein data bank) format. Additionally, the results should allow for feedback of the structure.
Tools of the Trade
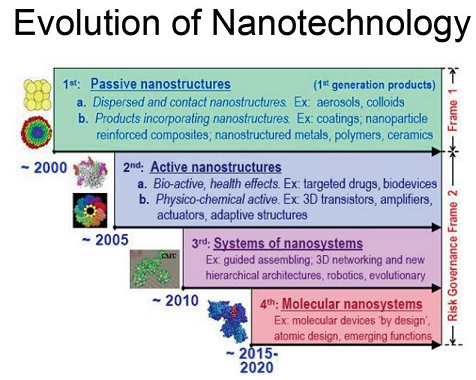
Why Synthetic DNA?Nature has already figured out how to assemble things that have high complexity. Up to now, nanostructures that have been invented are not complex and do not have the level of specificity that nature does. Synthetic DNA comes is important because it allows for this high level of specificity. Previous research has shown that 3D structures can be created using DNA. Thus synthetic DNA can be used to create structures of desired complexity and specificity. The work that we have done here improves the applicability of synthetic DNA structures to the field of materials science. By knowing the flexibilities of a structure, one can analyze its feasibility for the desired applications. Additionally, our method allows for feedback (explained further in the Results section) which allows a scientist to adjust the structure as needed for the application.
|