Biomod/2011/Harvard/HarvarDNAnos:Brainstorming
<html> <head>
<style>
- column-one { display:none; width:0px;}
.container{background-color: #f5f5f5; margin-top:50px} .OWWNBcpCurrentDateFilled {display: none;}
- content {width: 0px; margin: 0 auto auto 0; padding: 1em 1em 1em 1em; align: center;}
- column-content {width: 0px; float: left; margin: 0 0 0 0;padding: 0;}
.firstHeading {display:none; width:0px;}
- globalWrapper{width:1280px; margin:auto}
body {background: #F0F0F0 !important;}
- column-one {display:none; width:0px;background-color: #f0f0f0;}
- content{border:none;margin: 0 0 0 0; padding: 1em 1em 1em 1em; position: center; width: 800px;background-color: #f0f0f0; }
.container{ width: 800px; margin: auto; background-color: #f0f0f0; text-align:justify; font-family: helvetica, arial, sans-serif; color:#f0f0f0; margin-top:25px; }
- bodyContent{ width: 1267px; align: center; background-color: #f0f0f0;}
- column-content{width: 1280px;background-color: #f0f0f0;}
.firstHeading { display:none;width:0px;background-color: #f0f0f0;}
- header{position: center; width: 800px;background-color: #f0f0f0;}
- footer{position: center; width:1280px;}
</style>
</head> </html>
<html>
</html>
Early Meetings
2011-03-30: Meeting 1
Present: Adam, Sherrie, Nick, Shwinn, Evan
Pre-meeting Ideas
- Attaching a ssRNA with a known enzymatic function to the scaffold strand and seeing if the RNA retains its function as part of an origami structure
- Spell out our team name in origami
- This has been done (and see nanobreadboard idea below), but see DNA PAINT here http://dx.doi.org/10.1021/nl103427w -- in this case the readout of the name/barcode/pattern is optical, which is amazing since under normal circumstances the resolution of an optical microscope is coarser than the size of the entire DNA origami
- Making a "tight" cube from origami and attaching functional groups to the interior of the cube with the intent of creating a micro-environment where the functional groups could alter the pH relative to the surroundings
- Note the theme of how to design a truly good box or container from DNA
- Try using cations other than Magnesium to investigate their affect on reaction rate for multi-dimensional folding
- Making a robot with GFP attached to the eyes, and a mechanism that could allow him to move his arms (by say... altering the concentration of a particular staple sequence)
- Nanobreadboard
- See DNA PAINT idea above
- "Submarine" drug delivery
- Disease detection with DNA origami
- Some sort of primitive DNA calculator
- Connecting DNA self-assembly with research on self-assembly of macroscopic electrets
- Developing a procedure to detect and quantify free-radical damage of mitochondrial DNA.
- Catching natural RNA/DNA polymerase replication errors
- Very interesting idea -- see this paper www.mcb.harvard.edu/murray/publications/Elez_2010_20674359.pdf
- Using aptamers to detect the concentrations of various macromolecules, with the future application of providing real-time feedback on nutrient levels
- This is a very rich idea, especially in relation to the idea of circuits which implement "pattern classification" algorithms... see circuits idea below
- Implementing a sophisticated and easy-to-test DNA logic gate would be exciting
- Testing can often be done with http://en.wikipedia.org/wiki/Molecular_beacon molecular beacons or similar fluorescence based readout, e.g., in a fluorimeter
- You should check this out http://en.wikipedia.org/wiki/MAYA-II (a DNA computer that plays tic tack toe)
- Note the paper in nano-letters: http://dx.doi.org/10.1021%2Fnl0620684
- I would look forward to developing various algorithms to improve DNA origami self-assembly errors.
- We also need good ways to observe, quantify DNA origami assembly errors, direct nano-imaging (AFM or TEM) tells us if we have the right shape, but not necessarily how perfect it is at an atomic scale
- Antiviral drug: Since one subset of antiviral drugs uses antisense DNA/RNA to bind and shut off critical viral genes, maybe we can use a DNA origami box to transport these antisense molecules or use dynamic DNA structures to target the viral genome.
- Alternative energy: I read a Nature article that said someone from Arizona State is using DNA origami as a scaffold to hold an electron transport chain in place as a mimic of photosystem II. Maybe we can also exploit DNA origami's ability to serve as structural support to build something on top of it.
- Drug delivery: Put something in a DNA origami box. I'm not sure what that something would be. Like we talked about today, I'd be open to exploring box designs, like boxes that can keep certain things out.
- Investigate the relationship between staple lengths and DNA origami object stability and folding quality. The paper "A primer to scaffolded DNA origami" mentioned that this relationship can use some further study, so I jotted it down.
- No doubt about it: William Shih's lab has a very active research program on this question right now
- Electronics: This isn't really a thought-out idea; I just found it cool that IBM built a chip from DNA origami a couple years ago. I don't really know much about electronics, so I'm not sure what we could do along these lines.
- I think it would be cool to combine nanoscale control, metals and metal nano-particles, and fluorescence from small molecules -- metal enhanced fluorescence is amazing -- for instance, you can even make a fluorophore emit light in a SINGLE direction, basically making an antenna for light. You can also greatly enhance fluorescent signals for biomolecular detection applications.
Questions
- What is the difference between a Holliday junction and an antiparallel crossover created by an oligonucelotide staple?
- Why does the order of staples attaching to the scaffold not matter?
- How do expanded DNA alphabets work?
- How do walkers determine where to start?
- why bending within an origami structure happens
- why blunt-end stacking occurs.
- In caDNAno, how do we "draw" the structure that we want?
- After letting such a program run, does it "spit out" a list of staple sequences that one would then order from a third party?
- When using caDNAno or other similar programs, is it assumed that we are using the "known" sequence of the m13 bacteriophage as our scaffold strand?
- Is it possible to buy synthetic scaffold strands and input their sequences into such programs?
- Why is there such a heavy dependence on magnesium ions for folding? Is there another cation that we could substitute that may lead to a faster reaction rate (especially in the case of multi-dimensional origami structures)?
- Is there any previous research on RNA origami? If not, does the lack of such literature have anything to do with the ease of manipulating RNA (i.e., very rapid denaturation) or the cost of synthetic RNA?
- How does kinetic capillary electrophoresis control the rate and equilibrium constants of aptamer interactions?
- What are the current processing speeds of DNA logic gates?
- To what extent has DNA origami been shown to influence protein folding?
- I have read a paper in which a mouse antibody was able to specifically detect and bind to fullerenes. How well has in vivo nanoparticle-antibody recognition been elucidated?
- How do you find the best staple design in terms of sequence and length? Length constant, how much does the specific sequence of staple strands matter? Does Tm differ a lot by sequence?
- Under what circumstances do you use a honeycomb versus a square lattice? What advantages does a more tightly packed lattice (square) have over a less tightly packed one (honeycomb)?
- How are different DNA domains linked?
- How does twist occur in DNA origami structures?
- Has anyone ever tried a triangular lattice? Does that not work? Would the helices be too tightly packed?
Meeting Notes
- ~20 teams at Jamboree
- Clear, well-defined goal that fits time scale
- Narrow down to ~2 ideas by next meeting
- Obsessive notes on openwetware wiki!
- First day at lab: June 6, 9am, Wyss Institute
- Team name: HarvarDNAno
- 21 bases per P1 = 360 degree x 2 = 720 degrees. 720 degrees/3 = 240 degrees --> hexagonal. Crossover possible every 240 degrees = 7 base pairs.
- 8-60 base staples
- Short - don't stably bind
- Long - synthesis of sequence is not accurate
- Main themes:
- 1) DNA + X Hybrid Structures, where X = RNA, Aptamer, or Modified DNA
- 2) Boxes, boxes that exclude water
- 3) Circuits, logic gates, computing, dynamics, gene expression
2011-04-03: Informal Meeting 1
Present: Sherrie, Nick, Evan
Possible topics
- 1. DNA hybrid structures
- 2. Building special boxes
- 3. Gene expression and dynamics
- 4. Production of DNA nano-structures inside the cell
Some background
- Gene Expression
- DNA origami being used to detect RNA.
- Didn't find much else when I searched for DNA origami/dynamic DNA and gene expression.
- Why we might care to look into dynamic DNA and gene expression:
- 1) "For example, the hybridization chain reaction using RNA hairpins can function inside living cells and can even be used to selectively kill cancer cells via the protein kinase R pathway."
- "Dynamic DNA nanotechnology using strand-displacement reactions"
- 2) Antiviral applications
- 1) "For example, the hybridization chain reaction using RNA hairpins can function inside living cells and can even be used to selectively kill cancer cells via the protein kinase R pathway."
- DNA origami being used to detect RNA.
- From Adam
- Interesting findings from a fun blog by an eminent scientist, both relevant to us: "This has finally reached an apotheosis in which Somalogic, the latest incarnation of an aptamer company, has developed literally thousands of aptamers with modified nucleotides that bind to their targets in the low nM to pM range. (see, for example, Gold et al. (2010), PLoS One, 5:e15004).... Thus beginneth the Second Coming of Aptamers." See [1] and [2].
- Production of DNA nano-structures inside the cell (rather than trying to get them in from the outside, produce them inside)
- This is perhaps an "insanely difficult" bio-mod project -- i.e., to do something with both nano-structures and cells that is non-standard for the DNA nano-tech community -- but if you are really ambitious as a group and interested in this, I would be willing (and actually rather excited) to consider whether it is feasible to build on this idea
- One option is here: [3]
- Another is here: [4]
- I know the guy that did the 1st paper (he's in William Shih's lab). The second one might be harder, because I don't know if we can get our hands on the vector.
- I like ssDNA better than ssRNA since in-vitro prototyping is easier and less expensive.
- Also, we should do it in bacteria -- much easier / faster than mammalian cells for prototyping
2011-04-06: Meeting 2
Present: Nick, Sherrie, Shwinn, Evan, Adam
Ideas and Problems
Gene expression
- Single stranded DNA; RNA possible?
- Gene silencing
- Expression --> Triggered molecular event (e.g. silencing, optical signal)
Detection
- In vivo
- Related to gene expression
- In a test tube
- Detect color change, precipitate, fluorescence?
- With origami (output)
- With circuits (proteins/aptamers)
- Idea: a logical box with multiple locks and AND/OR gates
- Problem: how do we detect one molecule in an environment containing trillions of other ones?
- Molecular beacon: binding causes conformation change causing signal (fluorescence)
- Aptamer triggers exposure of DNA sequence
- Amplification
- Dave Zhang created DNA circuits that do enzyme-free, isothermal amplification: catalytic DNA circuit
- Idea: cascades, where each level lights up?
- Aptamers
- Time scale for producing evolved aptamers?
- Molecular beacon
- Binding -> conformational change -> fluorescence
- Fluorophore and quencher
- Questions:
- How big is a fluorophore?
- How many can we fit into a DNA origami box?
- How do we get them into a DNA origami box? How do we get them out?
Boxes
- Chemical properties
- What if we changed DNA at less than 1 nm scale? Seeman: B -> Z Transition
- Shawn Douglas's Box
- What would be the purpose of making a box?
- Idea: contains a LOT of fluorescent proteins that can be released all at once with lock and key mechanism
- Idea: catalytic boxes - boxes that when opened open other boxes, boxes that when opened opens other copies of itself
Produce DNA nanostructure in vivo
- Why?
- iGEM winners last year: class of Zinc fingers that have a combinatorial number of binding sequences, bind proteins to them --> synthetic metabolic pathways
- How?
- Shwinn wrote a paper on spectroscopic ways of detecting free radicals in cells
Capabilities
Scale
- 100 nm = max size of origami
- 10.5 bases = 1 turn in DNA
- 0.3 nm = 1 base
- 7 nm = 2 turns
- Precision = 1 nm
- Small molecule = less than 1 nm (scale of chemistry)
- 1-100 nm is scale of nanotechnology
- 100 nm-1 micron is scale of nanofabrication
- E. coli = 1 micron
- Proteins = several nm
Things to investigate
- Evan and Shwinn: catalytic circuits and amplification
- Nick: fluorophore attachment to DNA, types of boxes already constructed
- Sherrie: detection mechanisms in use today, their limits
2011-04-11: Informal Meeting 2
Present: Nick, Shwinn, Sherrie
Ideas to be discussed and investigated
Nick
- I am interested in developing a six-sided “logic” box that is held together by a series of and/or gates. These logic gates may be achieved using either strand displacement methods or aptamers. Ideally these gates will be opened in the presence of a certain identified viral sequence or toxin present. Application of such a logic box could, in theory, be utilized in or outside of the cell. However, I plan to focus on its application outside of the cell, inside the test tube. Potential applications for such technology include detecting toxins and/or viral agents in water. In addition, I hope to be able to attach fluorophores to the inside of our box that, upon activation of the logic gates, will open and fluoresce without the quenching effect of being stored close together inside of our DNA origami box. Before our next meeting, I plan to investigate how attachment of fluorophores to DNA may be achieved, and how to attach logic gates to our origami structure that will detect undesirable toxins.
Evan
- I am interested in constructing amplification pathways that can be used to detect minimal concentrations of a certain RNA or ssDNA strand (perhaps part of the HIV genome) in a test tube. The pathway might take form either as multiple levels of strand displacement or as a "self-opening" box. This box would come equipped with a lid, a lock-and-key mechanism, and an OR logic gate. This logic gate would accept either the RNA strand that we are detecting or a "key" strand. Each box, when closed, would contain a fluorescent protein, a quencher, and hundreds of the key strands; when opened, the fluorescent protein would be activated because of its loss of proximity with the quencher, and the key strands will be free to open more of the boxes. Both pathways could potentially be provided one day through a single, cheap "powder" that could be used to detect HIV in third-world countries where more technical methods involving PCR and enzymes would be impossible. I will try to put these two ideas on paper as diagrams/equations by Sunday. These ideas, especially the self-opening box, have a lot in common with Nick's proposal.
Sherrie
- I would like to explore how dynamic DNA interactions can contribute to the detection of a certain toxin or nucleic acid in vitro (later on, perhaps in vivo). Potentially, the presence of a certain molecule decreases the binding affinity of a region of dsDNA such that the DNA separates into two strands, one of which interacts with the molecule and the other of which is free to cause a cascade leading to macroscopic detection of the molecule. Such a mechanism can be integrated with Nick's box and Evan's amplification pathway. For our next meeting, I will look into current detection methods and their limitations, as well as how the characteristics of DNA might allow it to overcome these limitations (or, alternatively, create new obstacles). I will also compile a list of molecules 1) that have been relatively easy to detect in the past and may be similarly easy for us to detect with DNA, 2) that have been difficult to detect in the past and may be easier to detect with DNA, and 3) whose detection would prove meaningful to the human race. Applications include detection of gene expression in organisms and of heavy metals, parasites, and pathogens in water sources across the world.
Shwinn
- I would like to design an improvement over currently existing logic gates that can be modulated to detect various small molecules using specific sequences on aptazymes that can cleave a specific RNA sequence and will produce a certain output depending on the construction of the AND/OR/XOR gates. I would like to see whether this output can be amplified to a macroscopic signal. In developing a sound logic gate, we can hope to accurately measure combinatorial small molecule binding and detection within living systems.
Discussion Notes
Detecting heavy metals and bacteria
- Heavy metals already easily detected by relatively cheap test kits, DNA may not bring an advantage
- Bacteria detected by carbon nanotubes covered with aptamers
- Carbon nanotubes are biotoxic
- Replace carbon nanotubes with DNA origami and measure electrical conductivity?
Logic box
- Attach fluorophores to staple strands
- DNA tweezers logic gate?
- But not too much like Shawn's box
- A six-sided box "unzipping" along its edges?
- Multiple sides need to be unzipped by respective keys to open the box: AND/OR gates
- Random thought: What would DNA origami be like if we made stuff with A-DNA or Z-DNA?
- Probably can't use caDNAno anymore...
Presentation for Adam
- Nick: Attaching fluorophores to DNA origami
- Shwinn: Logic gates with DNA
- Sherrie: Detection of bacteria with aptamers
- Evan: Amplification
2011-04-17: Meeting 3
Present: Evan, Nick, Sherrie, Shwinn
The inspiration
Zhang et al. detection of catalyst
Need:
- 1) Substrate
- 2) Fuel
Add catalyst
- 2 hours --> maximum fluorescence 0.8 A.U
- Push fluorescence farther?
- How much catalyst? 10 nM
- How much fuel?
Our idea
Input
- Logic
Amplification
- Catalytic reaction cycle
- What are the limits?
- 1) Minimum detectable signal (smallest amount of catalyst): amount of amplification (> threshold), false positives (leaks that go above threshold)
- 2) Rate (linear, 1 output/cycle, 1*cycles*[catalyst]? exponential, n^cycles?)
Questions
- 1) What is maximum fluorescence that the system can product --> if we use the maximum concentration of substrate in practice?
- 2) How far off are we from a macroscopic result (fluorescing test tube)?
Boxes
- 2 boxes?
- 1 box with OR gate?
2011-06-10: Beginning of Summer!
- By next Thursday we're aiming for:
- a working prototype of sphereCAD, so that we can easily pattern the inside and outside of the sphere
- full sequences for the box design suggested by Tom, with the spring-loaded lid
- ordering cargo and associated strands
- To be posted on data section of wiki:
- caDNAno files for unedited sphere and disk
- staple-checking scripts
- 3D model of sphere
- sphereCAD software prototype
- caDNAno files for sphere with inner and outer attachments
- Illustrator drawings for Tom's design
- caDNAno file for Tom's design
- sequence output and additional sequences for Tom's design
- plan for work in the following 3 weeks
2011-06-16
- Evan plans to see if he can get his box to open and close using:
- Azobenzene
- Strand Displacement of Azobenzene-like lock
- Intra-molecular strand displacement
- Next, Evan plans to investigate attaching cargo inside of the box using:
- Azobenzene
- Strand displacement
- Di-sulfide linkers
- Nick and Sherrie plan to separate the sphere staples into the following categories:
- Matching strands on equator
- Matching strands not on equator
- Non-matching strands on equator - theirs
- Matching on equator - theirs
- Non-matching strands on equator - ours
- Matching on equator - ours
- Nick and Sherrie are then going to explore opening and attachment using a permutation of:
- Azobenzene
- Strand displacement
- Di-sulfide linkers
- Restriction Enzymes
Unexplored Project Ideas (for another summer!)
Origami Designs
Torus Design
- We want to explore the idea of working around weakness of sphere design (two holes at poles) by creating the same orientation of rings and additionally extending an inner layer that connects the top and bottom of the sphere to create the "no holes" torus
- Potential ideas:
- Thiolated strands within the "inner" layer of rings
- Strands exist as cross-linked disulfide bridges in natural confirmation
- Upon exposure to reducing environment, disulfide bridges cleaved to temporarily create small crevice within torus for nanoparticle uptake
- Time-sensitive re-oxidation to close torus
Concentric Rings
- We have replicated in caDNAno the Nine Concentric Rings structure in Han et al
Ellipsoid
- received caDNAno file from Han
Vase
- received caDNAno file from Han
Opening/Closing and Loading/Solubilization Mechanisms
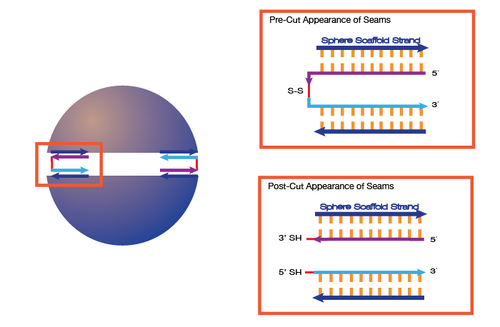
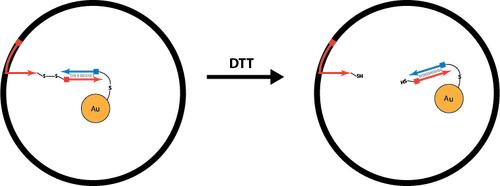
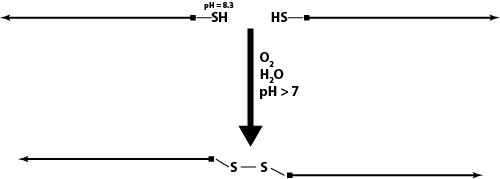
Disulfide Linkers
- How to Create Disulfide-Linked Staples
- Disulfide Linkers as a Mechanism to Open the Sphere
Advantages:
- cleaved by DTT or TCEP, which are small molecules that would quickly diffuse into the sphere
Experiments:
- We are currently testing our ability to create disulfide bonds by just using two 5'-thiolated strands
- We are trying to find the optimal pH for oxidation by using a pH reaction ladder and trying to determine if bringing pH back down will reverse the reaction
- We also want to test the cleavage of the disulfide bond with DTT
- We will analyze this system with PAGE
- NOTE: As of July 1st, we are no longer pursuing the use of disulfide linkers within our structures due to the nature of the cargo we wish to contain within our origami
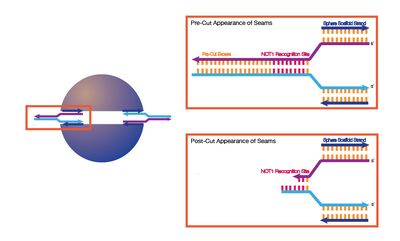
Restriction Enzymes
Advantages
- Precise cut locations
- m13mp18 does not contain the cut sequence of the NOT1 Restriction Enzyme
Disadvantages
- Not sure if this is entirely compatible with DNA origami, but have designed a mechanism outlined below to see if such a configuration will work well
- Not sure of how long pre and post-cut excesses should be
- Restriction Enzyme Model for Opening a Sphere
Additional Ideas
- Hetero-bifunctional crosslinkers that convert internal amine-modified bases to thiols
Cargo
Using Small 2D or 3D Origami as Cargo
- Triangle
- Nanoball
- Ring
<html> <a href="http://www2.clustrmaps.com/counter/maps.php?url=http://openwetware.org/wiki/Biomod/2011/Harvard/HarvarDNAnos:Brainstorming" id="clustrMapsLink"><img src="http://www2.clustrmaps.com/counter/index2.php?url=http://openwetware.org/wiki/Biomod/2011/Harvard/HarvarDNAnos:Brainstorming" style="border:0px;" alt="Locations of visitors to this page" title="Locations of visitors to this page" id="clustrMapsImg" /> </a> </html>