Beauchamp:CreateStndSurfModNew
Beauchamp Lab wiki
|
These steps are no longer required and may not be useful
for current instructions, please see
Older instructions that are obsolete
- First, we create an AC-PC aligned version of the surface, as follows
Find the AC-PC transform using the AFNI script @auto_tlrc which calls various other programs. The "-rigid_equiv" option computes a rigid transformation into TLRC space (this is the AC-PC alignment step without the stretching entailed by TLRC) The "-suffix none" adds the transformation to the existing dataset.
@auto_tlrc -suffix none -base TT_N27+tlrc -input {$ec}_SurfVol+orig -rigid_equiv
- Run ConvertSurface to translate the original space FreeSurfer surfaces according to the transformation matrix from the first step (note that this converts the filetype to *.ply). This makes ACPC-aligned versions of various version of the surface. We don't do all of them because they are rarely used (but we could).
ConvertSurface -i_fs lh.smoothwm.asc -sv {$ec}_SurfVol+orig -o_ply lh.smoothwm_tlrc -tlrc
ConvertSurface -i_fs lh.inflated.asc -sv {$ec}_SurfVol+orig -o_ply lh.inflated_tlrc -tlrc
ConvertSurface -i_fs lh.pial.asc -sv {$ec}_SurfVol+orig -o_ply lh.pial_tlrc -tlrc
ConvertSurface -i_fs lh.sphere.asc -sv {$ec}_SurfVol+orig -o_ply lh.sphere_tlrc -tlrc
ConvertSurface -i_fs rh.smoothwm.asc -sv {$ec}_SurfVol+orig -o_ply rh.smoothwm_tlrc -tlrc
ConvertSurface -i_fs rh.inflated.asc -sv {$ec}_SurfVol+orig -o_ply rh.inflated_tlrc -tlrc
ConvertSurface -i_fs rh.pial.asc -sv {$ec}_SurfVol+orig -o_ply rh.pial_tlrc -tlrc
ConvertSurface -i_fs rh.sphere.asc -sv {$ec}_SurfVol+orig -o_ply rh.sphere_tlrc -tlrc
- Create spec files to load these into SUMA
cp /Volumes/data9/surfaces/scripts/both_tlrc.spec . cp /Volumes/data9/surfaces/scripts/lh_tlrc.spec . cp /Volumes/data9/surfaces/scripts/rh_tlrc.spec .
Check with
afni &
suma -spec both_tlrc.spec -sv {$ec}_SurfVol+orig
Make sure to click "Talairach View" after AFNI starts to see the correct surface to volume correspondence.
- Run MapIcosahedron to create a standardized surface with EXACTLY 156,252 nodes.
Because each surface now has the same number of nodes, we can average at each node across subjects. Note that the program adds "_std" to the given prefix. We used to use an additional smoothing step to normalize the size of the notes (argument -it 20) but according to Ziad this is no longer needed.
MapIcosahedron -overwrite -spec lh_tlrc.spec -prefix std_ -morph sphere -ld 125 & MapIcosahedron -overwrite -spec rh_tlrc.spec -prefix std_ -morph sphere -ld 125
MapIcosahedron creates its own SPEC files but it is easier to use a single file that contains both hemispheres
cp /Volumes/data9/surfaces/scripts/both_tlrc_std.spec .
Check with
afni &
suma -spec both_tlrc_std.spec -sv {$ec}_SurfVol+orig
Make sure to click "Talairach View" after AFNI starts to see the correct surface to volume correspondence.
- Register the standardized surface with the experiment data so that we can map functional data onto the standardized surface. Because the "+tlrc" surface volume is not warped, it can be registered to the original anatomy, as follows:
@SUMA_AlignToExperiment -prefix {$ec}_SurfVol_std_Alnd_Exp -ok_change_view -exp_anat {$ec}anatavgSS+orig -surf_anat {$ec}_SurfVol+tlrc
This produces an "+orig" view dataset which allows all functional data to be viewed normally. Check with
afni &
suma -spec both_tlrc_std.spec -sv {$ec}_SurfVol_std_Alnd_Exp+orig
- The most common use of standardized surfaces is to make group average datasets. In order to do this, the functional data must be mapped to the surface. This must be done separately for each hemisphere.
foreach hemi (rh lh)
3dVol2Surf -spec std_{$hemi}_tlrc.spec -surf_A smoothwm -surf_B pial -sv {$ec}_SurfVol_std_Alnd_Exp+orig \
-grid_parent {$ec}v1mr+orig -map_func ave -f_steps 15 -f_index nodes -out_niml {$ec}v1mr_{$hemi}.niml.dset
end
There is an infinite variety of mapping functions to use.
A very simple way to is to take voxels that intersect a surface and give each node the value of the voxel that it intersects.
This is similar to the way the AFNI and SUMA GUIs communicate. This is a good way for testing out different mapping functions (compare with the internal function).
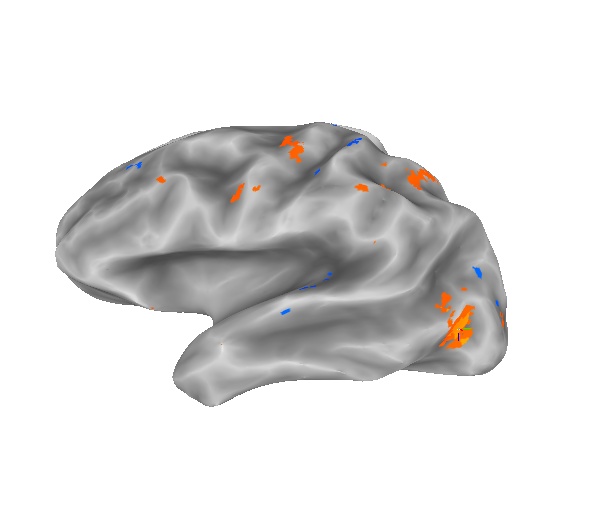
This picture shows the results of the built-in mapping function (all t>5 for subBRIK#2 of FLv1mr+orig).
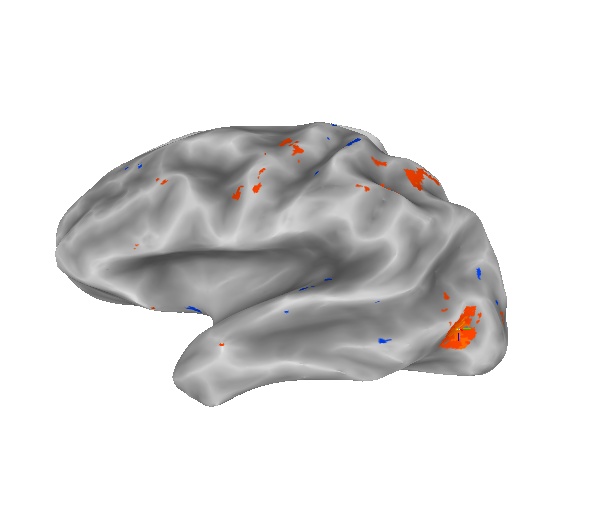 The next pictures shows the results of the following command.
The next pictures shows the results of the following command.
set sd = /Volumes/data9/surfaces/graham_tim/FL/SUMA/
set sb = 2
3dVol2Surf -overwrite -spec {$sd}/{$ec}_{$hemi}.spec -surf_A smoothwm -sv {$ec}_SurfVol_Alnd_Exp+orig \
-grid_parent {$ec}v1mr+orig'['{$sb}']' -map_func mask -f_index nodes -out_niml {$ec}v1mr_{$hemi}_sb{$sb}_mask.niml.dset
If could be argued that voxels in the gray matter might not intersect the smoothwm surface. So, a mapping function that takes the average of all voxels between the smoothwm and pial surfaces might be more accurate. This pictures shows the results of such an average mapping function.
3dVol2Surf -spec std_{$hemi}_tlrc.spec -surf_A smoothwm -surf_B pial -sv {$ec}_SurfVol_std_Alnd_Exp+orig \
-grid_parent {$ec}v1mr+orig -map_func ave -f_steps 15 -f_index nodes -out_niml {$ec}v1mr_{$hemi}.niml.dset
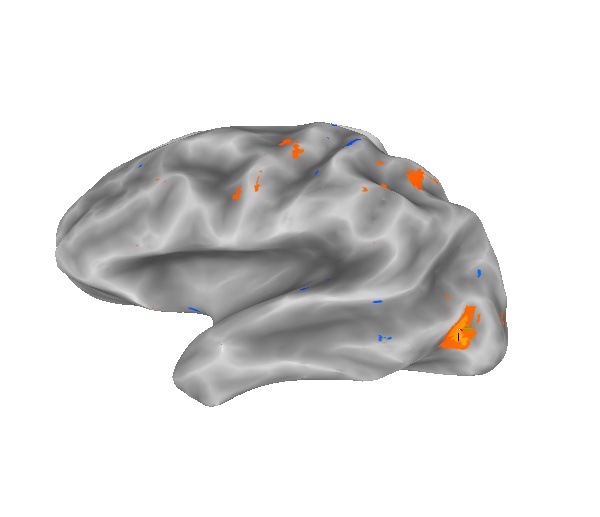 Because there is not a giant difference in the results between the different mapping functions, usually the simplest is preferred.
By default, a node that does not touch a voxel is assigned no value. For statistical analyses across subjects, it is best if every node has a value so that a mean and SD across subjects can be calculated. The simplest is to assign "0" to a node where there is no value (this is not actually correct, so it must be remembered that the lack of activation in a region such as the bottom or top of the brain e.g. ventral temporal cortex or motor cortex may in fact reflect the lack of data from this brain region). This is done with the "oob_value" parameter.
Because there is not a giant difference in the results between the different mapping functions, usually the simplest is preferred.
By default, a node that does not touch a voxel is assigned no value. For statistical analyses across subjects, it is best if every node has a value so that a mean and SD across subjects can be calculated. The simplest is to assign "0" to a node where there is no value (this is not actually correct, so it must be remembered that the lack of activation in a region such as the bottom or top of the brain e.g. ventral temporal cortex or motor cortex may in fact reflect the lack of data from this brain region). This is done with the "oob_value" parameter.
3dVol2Surf -overwrite -spec {$sd}/{$ec}_{$hemi}.spec -surf_A smoothwm -sv {$ec}_SurfVol_Alnd_Exp+orig -oob_value 0 \
-grid_parent {$ec}v1mr+orig'['{$sb}']' -map_func mask -f_index nodes -out_niml {$ec}v1mr_{$hemi}_{$sblabel}.niml.dset
- Other uses for the standard surface are to plot co-ordinates (e.g. electrode locations) from individual subjects on a single brain.
To plot 3-D coordinates to the standardized surface, first we convert them to AC-PC space
Vecwarp
and then we map to the surface
SurfaceMetrics
- To create sulcal depth files on the standardized surface, we map the existing sulcal depth file onto the standardized surface. It can then be loaded in with the SUMA surface controller for visualization.
SurfToSurf -prefix test.1D -output_params Data -i lh_acpc_std.smoothwm.asc -i lh.smoothwm.asc -data lh.sulc.asc
If a sulc file does not exist already it might be possible to create another one with these steps
3dSkullStrip -input TT_N27_SurfVol+tlrc.HEAD -o_ply TT_N27_BrainSurface.ply SurfToSurf -i lh_acpc_std.smoothwm.asc -i TT_N27_BrainSurface.ply -prefix newdepth.1D -output_params DistanceToSurf
But this fails because the Surfaces are not in alignment.
An older version of this page is at Creating Standardized Surface Models