Beauchamp:SurfaceAveraging
Return to Overview of Cortical Surface Models
Creating Surface Averages of Functional Data
Averaging on the surface has many advantages over averaging in the volume but requires more processing steps. What is needed for each subject is a surface that is folded and aligned to the individual subject anatomy but that has the same number of nodes in each subject (this way a node-wise group map can be created) AND has the nodes aligned to a template brain (this way any given node has the same anatomical location in each subject). Each subject's functional data is mapped to that subject's standardized surface and then node-based group maps are created. The process must be performed separately for the left and right hemispheres.
SUMA automatically creates surfaces that have the desired properties, the std.141 brains. First, we transfer the functional data from each subject to the standard brain with a command line like this:
set s = OB
set sd = /Volumes/data/BCM/{$s}/fs/SUMA
set fd = /Volumes/data/BCM/{$s}/AFNI
cd $fd
3dVol2Surf -overwrite -spec {$sd}/std.141.fs_both.spec -surf_A std.141.lh.smoothwm.gii -sv fs_SurfVol_Alnd_Exp+orig -oob_value 0 \
-grid_parent "stats.OB2+orig[MouthFull#0_Coef,EyeFull#0_Coef]" -map_func mask -out_niml {$sd}/{$s}_EM_lh.niml.dset
Although surface registration between subjects is more accurate than volume registration, it is still good to spatially smooth the functional data to account for anatomical variability. It is best to only smooth once, on the surface, so remove any other smoothing steps in the preprocessing. 5 mm is a reasonable amount of smoothing; experiment with smaller amounts.
SurfSmooth -met HEAT_07 -fwhm 5 -input {$gd}/{$s}_lh_EM.niml.dset -i {$sd}/std.141.lh.smoothwm.gii -output {$gd}/{$s}_lh_sm_EM.niml.dset
After creating surface functional files for each subject, we can use AFNI's group analysis tools exactly as with volume datasets. For instance,
3dttest++ -overwrite -paired -prefix lh_sm5mm_MvsE.niml.dset \ -setA MS_lh_sm_EM.niml.dset'[0]' MU_lh_sm_EM.niml.dset'[0]' MX_lh_sm_EM.niml.dset'[0]' \ -setB MS_lh_sm_EM.niml.dset'[1]' MU_lh_sm_EM.niml.dset'[1]' MX_lh_sm_EM.niml.dset'[1]'
or
3dANOVA3 -type 4 -alevels 2 -blevels 2 -clevels 20 \ -dset 1 1 1 MS_lh_sm_EM.niml.dset'[0]' \ -dset 1 1 2 MT_lh_sm_EM.niml.dset'[0]' \ ... -dset 2 2 20 OB_lh_sm_EM.niml.dset'[3]' \ -fa MouthEye -fb FullMasked -fab MEvsFullMasked -adiff 1 2 MouthvsEye -bdiff 1 2 FullvsMasked -bucket ANOVA_lh_v1
Displaying the Average Functional Data
After it is averaged on the surface, the fMRI data can be displayed on an individual participant brain or on average brain (see next section for instructions on creating an average anatomical surface). To display on an individual, any std.141 brain can be used. For instance, to display on the Colin N27 brain,
set workdir = /Volumes/data/BCM/group_eyemouth_ISV
Set this to the directory where your group average functional data live.
cd /Volumes/data/BCM/N27 cp @ec_surf std.141.MNI_N27_both.spec *std.141*gii *std.141*sulc* *std.141*2009* $workdir
Copy the N27 surfaces over and use to display results.
Creating Averaged Anatomical Surfaces
To get an idea of how well or poorly the surface normalization works, it can be nice to create an average anatomical surface by taking the mean position in 3D space of each individual node. Here are the steps for creating averaged surfaces from anatomical data.
Convert each subject's surface to a 1D file using ConvertSurface:
foreach s ( $ss )
echo $s
set sd = /Volumes/data/{$g}/{$s}/fs/SUMA
ConvertSurface -overwrite -i {$sd}/std.141.lh.smoothwm.gii -o_1D {$s}_lh {$s}_lh
end
Average the location of each node on the standardized surface using 3dMean
3dMean -prefix AvgSurf_lh ??_lh.1D.coord
Convert the average locations back into a surface file that we can load it into SUMA
ConvertSurface -i_1D AvgSurf_lh.1D OB.1D.topo -o_gii AvgSurf_lh.gii
Load into SUMA with
suma -i AvgSurf_lh.gii
Here are some images illustrating this process.
Single subject surface model  Average surface model across 20 subjects. Note that it is smoother than the individual subject.
Average surface model across 20 subjects. Note that it is smoother than the individual subject.  To quantify regional differences in anatomical variability, we can calculate the standard deviation across subjects at each node (similar to average difference between two surfaces above) using
To quantify regional differences in anatomical variability, we can calculate the standard deviation across subjects at each node (similar to average difference between two surfaces above) using
3dMean -stdev -prefix AvgSurf_lh_v1_SD ??_lh.1D.coord
Plotted on the surface, lateral and medial views 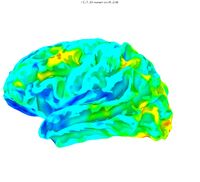

As expected, SD is low in regions that are consistent across subjects (such as the Sylvian fissure ) and SD is high in regions of anatomical variability (such as occipital pole). If desired, SurfSmooth can be used to partially inflate the averaged surface for easier visualization.
Obsolete Stuff
This method supersedes (by being simpler and better) the method described in Argall, B. D., Saad, Z. S., and Beauchamp, M. S.: Simplified intersubject averaging on the cortical surface using SUMA. Hum Brain Mapp 27:14-27, 2006. The previous method is described on obsolete web pages: