BME100 s2017:Group5 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
OpenPCR program HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25
FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
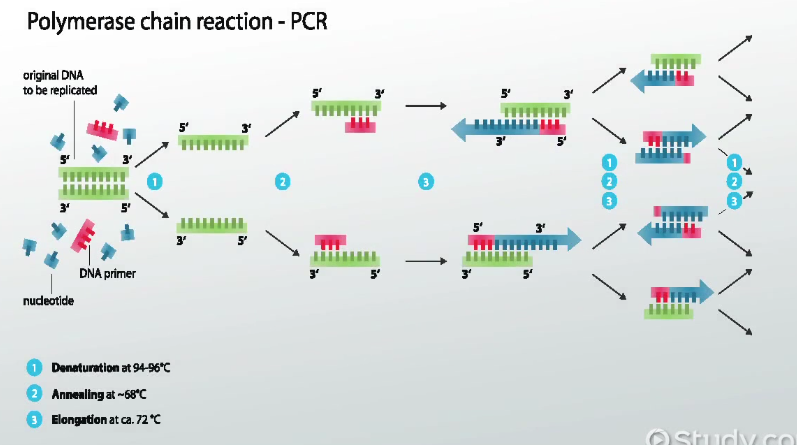
Research and DevelopmentPCR - The Underlying Technology The components of PCRPCR begins with the template DNA, which serves as a template or guide for the production of multiple DNA copies. Short pieces of DNA that are designed to be a DNA synthesis starting point, primers, are then introduced to catalyze the DNA replication process. Taq Polymerase, complex proteins that copy a cell's DNA before it divides in two, are then used to replicate the DNA since it is thermostable, meaning it won't denature when the sample is heated to high temperatures. Deoxyribonucleotides, or dNTP's, attach themselves to the end of a primer as the building blocks of DNA molecules to create the new single strand. Since the process continues again from the beginning, these are the strands that will then also go through PCR. Using Thermal Cycling in PCRThe process begins with the sample being heating at 95°C for 3 minutes, and the sample is then denatured at 95°C for 30 seconds. When denaturing, the DNA double helix separates to create two single stranded DNA molecules. It is then annealed at 57°C for 30 seconds. During this step, the primers base pair with the DNA strands. Single stranded DNA molecules naturally attempt to pair up, so primers attach to DNA before strands can rejoin. Next, the sample is extended at 72°C for 30 seconds which activates DNA polymerase. The DNA polymerase is activated and begins to add the complementary nucleotides on the DNA strand. It continues until it gets to the end of the strand and falls off. Base-pairing occurs during annealing and extending. During the bonding of base pairs, Adenine anneals exclusively to Thymine and vice versa, and Cytosine anneals to Guanine and vice versa. The final step keeps the sample at 72°C for 3 minutes, and the sample is cooled to 4°C for a final hold. Primers Binding to the Cancer DNA Template and Taq Polymerase amplifying DNA"Taq Polymerase: Definition & Function - Video & Lesson Transcript." Study.com. N.p., n.d. Web. 16 Mar. 2017. http://study.com/academy/lesson/taq-polymerase-definition-function-quiz.html
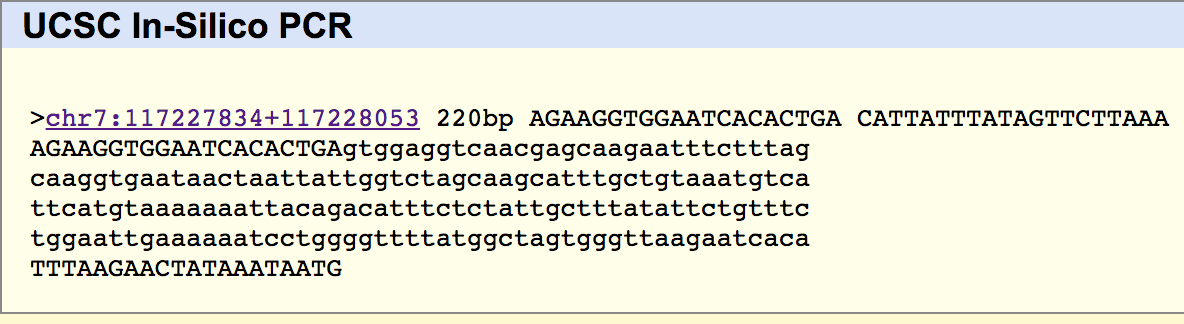
SNP Information & Primer DesignBackground: About the Disease SNP The SNP that was looked at was rs121908757. This pathogenic SNP is linked to the CFTR gene and is located on chromosome 7 and is located at numerical position 117587799. CFTR (Cystic Fibrosis Transmembrane Conductance Regulator) is a membrane protein and chloride channel that is encoded by the CFTR gene. This channel conducts chloride and thiocyanate ions across epithelial cell membranes. In the rs121908757 polymorphism, the CGT codon exists because AGT is changed to CGT. Primer Design and Testing Our group designed the primers by observing the code near the SNP. The non-diseased and diseased primers (pictured below) were input into the UCSC database and matched it against known sequences. The non-diseased primer had a match on the proper chromosome (7), but it was located in a slightly different position than expected. This is likely due to the different updating rate of the two databases that we used. The initial testing was done with the UCSC database from 2009, and when using the database from 2013, the base pair location was closer to the expected value. The diseased primer had no matches, which makes sense as the diseased gene is not what was databased. | ||||||||||||||||||||||||||||||||||