BME100 s2017:Group4 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
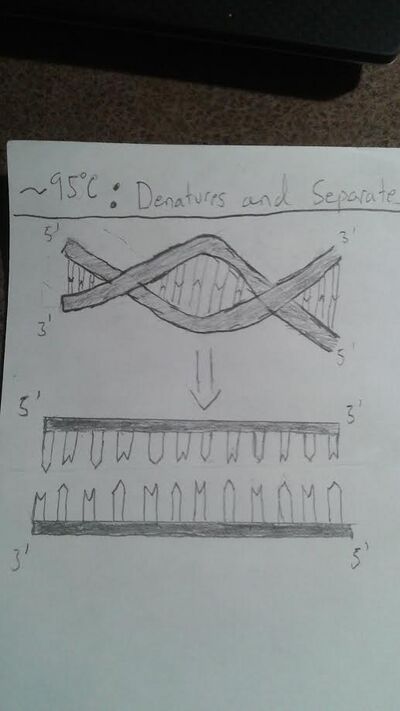
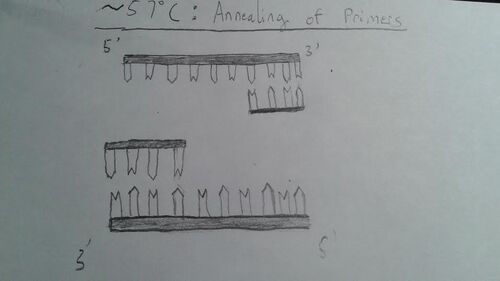
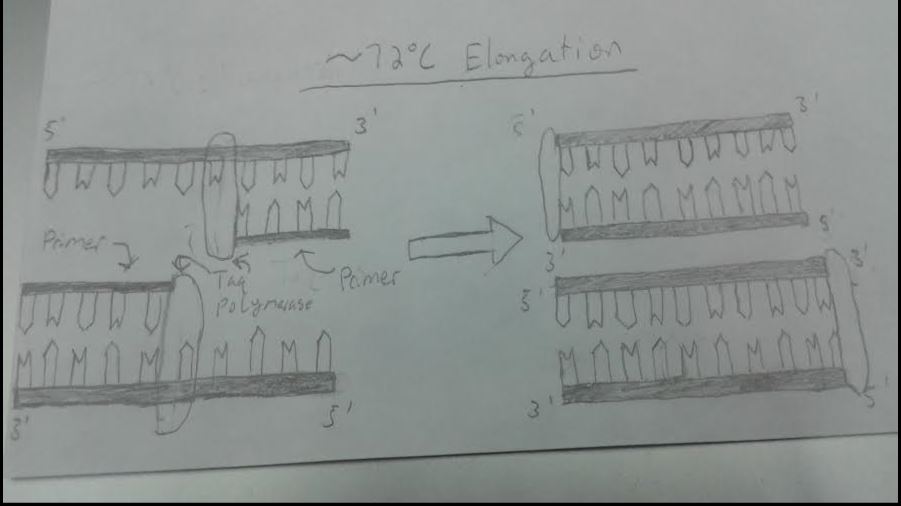
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes (to allow the DNA to denature and separate as much as possible) NUMBER OF CYCLES: 25 Denature at 95°C for 30 seconds (to allow the DNA to uncoil and the strands to separate) Anneal at 57°C for 30 seconds (to allow primers to attach to the DNA at a certain spot) Extend at 72°C for 30 seconds (to allow DNA polymerase to extend the desired portion of the DNA strand) FINAL STEP: 72°C for 2 minutes (to ensure that the DNA has been fully elongated) FINAL HOLD: 4°C (to let the DNA, in its fully replicated state, coil back together and act like DNA naturally would as well as to hold further replication)
Research and DevelopmentPCR - The Underlying Technology Q1: What is the function of each component of a PCR reaction?
Q2: What happens to the components (listed above) during each step of thermal cycling?
Q3: DNA is made up of four types of molecules called nucleotides, designated as A, T, C and G. Base-pairing, driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each base listed below? If you need help, use the “Build a DNA Molecule” tool at http://learn.genetics.utah.edu/content/begin/dna/builddna/
Q4: During which two steps of thermal cycling does base-pairing occur? Explain your answers Base-pairing occurs during annealing and elongation. During annealing, the primers will anneal to the target DNA; they have been designed to bind to a specific region of the DNA. Its bases will pair with the bases in the target DNA segment. During elongation, TAQ Polymerase will extend the primers by adding dNTPs to the target strand and make double stranded DNA.
At around 95 degrees Celsius, the hydrogen bonds between the AT and GC pairings are broken, which allows the template DNA to unzip and separate.
At around 57 degrees Celsius, 2 primers anneal to the desired region in the template DNA. They attach to the 3' ends of the top and bottom strand because DNA polymerase always builds 5' to 3'.
At around 72 degrees Celsius, the DNA polymerase becomes active and attaches the complementary nucleotide bases until it reaches the end. Taq polymerase is used in particular because it operates well at high temperatures.
After elongation occurs, the whole denaturing, annealing, and elongating process repeats itself for around 25 cycles in this case. After the third cycle, DNA segments that are purely the desired regions begin to appear. After all the cycles, millions to over a billion copies(depending on the number of cycles) of the desired DNA region will have been replicated.
SNP Information & Primer DesignBackground: About the Disease SNP A SNP is a single nucleotide polymorphism, which means that one nucleotide is replaced with another(the standard nucleotide is replaced with a different one due to a point mutation). In the specific case of this disease SNP, an adenine nucleotide is replaced with a cytosine instead. The allele changes from AGT to CGT when this SNP occurs. Although the majority of SNPs do not have negative disease related consequences, this specific SNP is pathogenic. In particular, it is linked with Cystic Fibrosis and is found on chromosome 7:117587799 in Homo sapiens(humans).
General Background Information: What is a nucleotide? A nucleotide is a compound consisting of a nucleoside linked to a phosphate group. The nucleotides form the basic structural unit of nucleic acids such as DNA. What is a polymorphism? A polymorphism is the condition of multiple forms of a single gene.
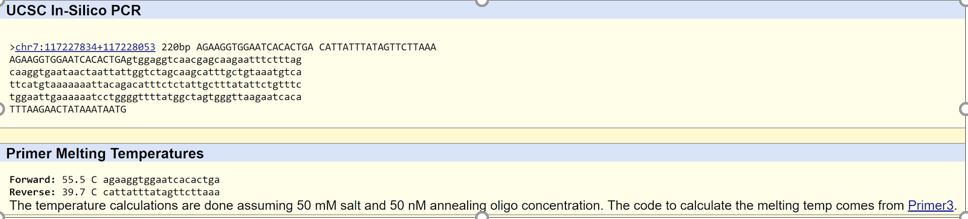
Primer Design and Testing
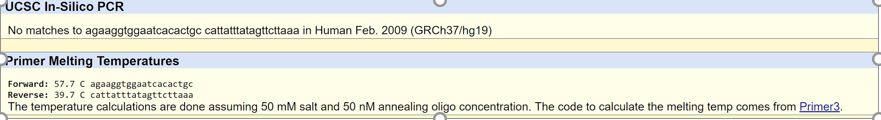
There was no result for the disease positive allele. This is because it is a mutation, so it is not part of the normal human genome. While this mutation occurs, it does not happen to the average human, so it was unable to be found in the database.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||