BME100 s2017:Group2 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
DNA Sample Set-up Procedure
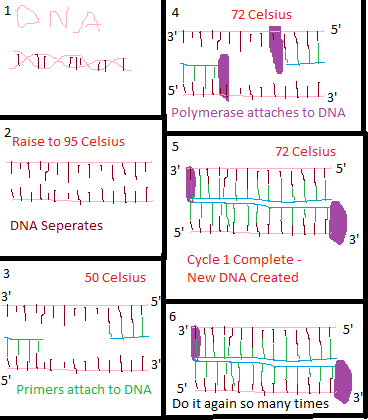
Heated Lid: 100°C (or 212°F) Research and DevelopmentPCR - The Underlying Technology The template DNA is the start point of the PCR process, it is what you are attempting to replicate, and is the base of the process. The primers are added, and are used to choose the specific area of DNA you are attempting to replicate. They target specific areas on the DNA, and attach to these areas to act as starting points for the replication. The DNA polymerase is then added; this is what actually builds the replicated strands of DNA by reading the DNA and attaching the appropriate nucleotides in the appropriate order. The dNTP’s are what actually make up the new DNA strand - these are the building blocks of DNA - adenine, guanine, cytosine, and thymine, and are matched by the polymerase to the original DNA strand to mirror the DNA. The initial step, raising the temperature to 95℃ for 3 minutes serves to separate the DNA double helix. This is known as denture and creates two single-strand DNA codes over 30 seconds. After this, the temperature is lowered to 57℃ for 30 seconds which allows the primers to sneak in between the DNA strands and lock onto the target areas. This is the anneal. The temperature is then raised to 72℃ (extend) for 30 seconds which allows the polymerase to lock onto the primers and begin creation of the new, replicated strand of DNA. The temperature remains here for 3 minutes which then allows the polymerase to do its’ work of creating the new DNA strands. The temperature is then held at 4℃ for extraction and preservation of the new DNA During this process, the adenine anneals to thymine and the cytosine anneals to guanine. The base-pairing itself occurs during the annealing phase, when the primer first locks onto the target section that matches the nucleotides it holds. It also occurs during the final extension phase where the polymerase creates a new strand of DNA by base-pairing the DNA strand it is attached to.
SNP Information & Primer DesignBackground: About the Disease SNP
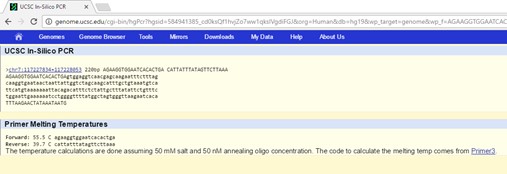
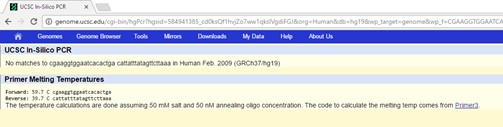
The results of the non-disease specific primer test resulted in base pair measurement of 220 base pairs. This is accurate because the forward primer was twenty base pairs long and the reverse primer was 200 base pairs away from the SNP position. Since the reverse primer reads from the 5’ direction to the 3’ direction from right to left, the entire number of base pairs from is equal to 220. Results from the disease -specific primer: The results of the disease specific primer were inconclusive due to the missense mutation which causes the change of the SNP rs121908757. Since the disease specific primer is based off of the change in the nucleotide of the original sequence, it will not detect any base pair amount because the point mutation is not present in the database for the CFTR gene. | ||||||||||||||||||||||||||||||||||