BME100 s2014:W Group9 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||
|
Xtreme DNA
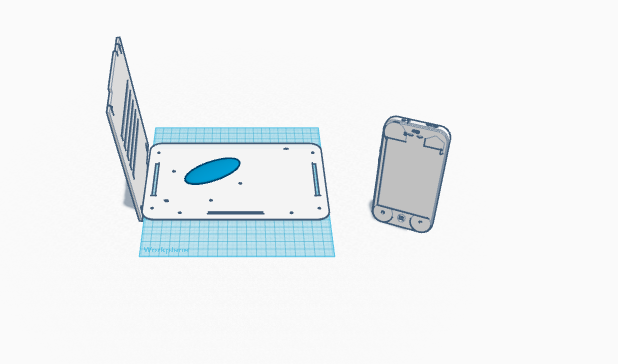
LAB 6 WRITE-UPComputer-Aided DesignTinkerCAD TinkerCAD is a program used to create a 3-dimensional design of an object or invention. It allows you to contort, change and alter the shapes in the free space provided. We utilized it, with pre-generated parts of our machine, to create a new and more effective design. Usually, the physical model created is then manufactured. In our case, we used the different parts of the OpenPCR machine to make a more effective and user-friendly machine.
Feature 1: Disease SNP-Specific PrimersBackground on the cancer-associated mutation The single nucleotide polymorphism rs237025 is found in the human chromosome 6:149721690. A sequence of alleles is changed from 'GTG' to 'ATG'. This variation occurs in the gene SUM04. This gene is responsible for encoding ubiquitin-related modifiers. These modifiers attach themselves to proteins to control activity, stability, and/or localization. Changing just that one small allele can cause effects with rheumatoid arthritis or type 1 diabetes.
Primer design
Feature 2: Consumables KitOur consumables kit will come in an organized container in sections that are clearly labeled as opposed to the original design which can cause clutter and confusion. It will include the plastic tip cartridges, a redesigned pipettor, and the necessary reagents to begin the experiment right away. The redesigned pipettor will now allow quicker pipetting of the reagents. It will remove the need of reattaching each individual plastic tip. Instead, a cartridge will be loaded into the pipettor and each time the plastic tip is ejected, a new tip will automatically discharged and become ready to use. This tip will also be sterile, preventing any type of error while using the OpenPCR machine.
Feature 3: Hardware - PCR Machine & FluorimeterThe OpenPCR machine and fluorimeter will come in a clearly labeled package side-by-side with the consumables. The slides will be packaged in a way that will prevent breaking during transport. All the necessary equipment will be included to begin immediately. Clear instructions will also be provided. The redesigned PCR machine will now include bluetooth capability, allowing for the removal of a transfer cable between the machine and the computer. Bluetooth is being used instead of Wi-Fi in order to prevent any connectivity issues. Instead, a smartphone app will be utilized in order to provide an accurate visualization of the recorded data. Within our packaging, we will provide a QR code that will allow for easy and quick installation of the application. This extensive application will include a self-timer camera and imageJ software. This will allow for mobile analysis of DNA.
Bonus Opportunity: What Bayesian Stats Imply About The BME100 Diagnostic Approach[Instructions: This section is OPTIONAL, and will get bonus points if answered thoroughly and correctly. Here is a chance to flex some intellectual muscle. In your own words, discuss what the results for calculations 3 and 4 imply about the reliability of PCR for predicting the disease. Please do NOT type the actual numerical values here. Just refer to them as being "less than one" or "very small." The instructors will ask you to submit your actual calculations via a Blackboard quiz. We are doing so for the sake of academic integrity and to curb any temptation to cheat.]
| ||