|
OUR TEAM
Mohammed Mousa, Maryam Ibrahim, Quintin Woods, Kayla Cummins, Kyndal Sorenson
LAB 4 WRITE-UP
Initial Machine Testing
The Original Design
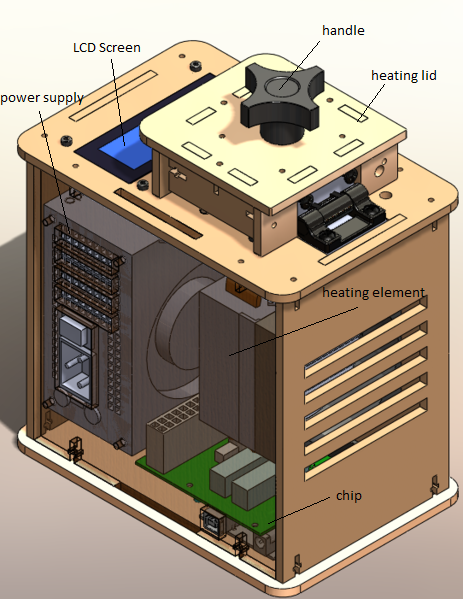
The PCR machine consists of a wooden box containing a heating element, power supply, and fan. On top of the box is the heating lid with handle. When the lid is lifted, slots for reaction tubes can be accessed. The LCD screen displays the current temperature, number of cycles completed, and estimated time remaining. The PCR machine is connected to a computer via USB. Once connected, Open PCR software is used to det the heating and cycling parameters. The computer monitor displays the same information as the LCD screen and gives an option to stop the experiment.
Experimenting With the Connections
When we unplugged the LCD screen from the chip, the screen turned off.
When we unplugged the white wire that connects the chip to the heating element, the machine was no longer accurate in its temperature readings.
Test Run
(3/19/14)
The machine had difficulty connecting with the computer software at first, but after a couple of tries and a new USB slot the issue was resolved. At first it ran slowly but was able to run through 11 cycles by the end of class. The machine passed.
Protocols
Thermal Cycler Program
HEATED LID: 100°C
INITIAL STEP: 95°C for 2 minutes
NUMBER OF CYCLES: 30
DENATURE: 95°C for 30 seconds
ANNEAL: 57°C for 30 seconds
EXTEND: 72°C for 30 seconds
FINAL STEP: 72°C for 2 minutes
FINAL HOLD: 4°C
DNA Sample Set-up
DNA Sample Set-up Procedure
- 8 reaction tubes were provided to us, each containing 50μL of PCR reaction mix.
- All reaction tubes were labeled according to the table above in order to avoid swapped results.
- 50μL of each DNA sample Mix was added to the correspondingly labeled reaction tube (using a new pipette tip for each transfer in order to avoid cross-contamination between samples).
- The reaction tubes were placed into the thermocycler. The thermocycler program detailed above was run so that PCR would occur in each reaction tube.
PCR Reaction Mix
- What is in the PCR reaction mix?
1) Taq DNA polymerase
2) dNTP's
3) MgCl2
DNA/ primer mix
- What is in the DNA/ primer mix?
1) Extracted sample of a particular patient's DNA
2) Forward Primer
3) Reverse Primer
Research and Development
PCR - The Underlying Technology
Listed below are the components of a PCR reaction and their functions
Template DNA
Serves as the template that allows for the entire reaction to occur. Without this, the designed primers wouldn't be able to bind to anything and as a result TAQ polymerase wouldn't be able to do combine the dNTPs to generate a large growth in DNA strands.
Primers
These are required to bind to the template DNA. These in a sense serve as a signal for the TAQ polymerase for where to start constructing a complimentary strand of DNA. These are pre-created to bind only to a specific sequence of DNA, the cancerous one in this case. A forward and a reverse primer are both needed because DNA polymerase can only work in one direction, so the same sequence must be made in opposite directions in order to satisfy the fact that DNA is composed of two strands: a leading and lagging strand.
Taq Polymerase
This is the enzyme that generates the new sequence of DNA using an original strand of DNA as a template. It binds to the primers, and collects the dNTPs that are floating around in the mixture to bind these complimentary to the given strand until the desired sequence is generated.
Deoxyribonucleotides (dNTP’s)
These are unpaired nucleotides that are floating around in the entire mixture. These are collected by the TAQ polymerase and form the nucleotide sequence that is complimentary to the template DNA sequence.
The steps in which PCR occurs
Initial Step: 95 degrees C for 3 minutes
In the first step of PCR where the samples were heated to 95ºC for three minutes, the two strands of DNA were separated from one another due to the high amount of heat in the reaction. This temperature and the amount of time that the samples were heated were both necessary because they allowed for an adequate amount of energy necessary to break the hydrogen bonds between the parallel nucleotides. This caused the DNA sequence to unzip, which opens up the opportunity for the entire reaction to occur.
Denature at 95°C for 30 seconds
The DNA was separated in order for the individual components to begin mixing together.
Anneal at 57°C for 30 seconds
The samples were cooled down, which allows for the forward and reverse primers to bind to their respective regions.
Extend at 72°C for 30 seconds
Causes the Taq polymerase which is catalyzed by the magnesium chloride to attach the buoyant dNTP’s to their complementary base pair.
FINAL STEP: 72°C for 3 minutes
The temperature is adequate enough to where the polymerase can generate a new strand of DNA from the template quickly without denaturing
FINAL HOLD: 4°C
The final step of the experiment. Ensures the test was carried out successfully and the machine stops running.
Base Pairings:
| Adenine (A) |
Thymine (T)
|
| Thymine (T) |
Adenine (A)
|
| Cytosine (C) |
Guanine (G)
|
| Guanine (G) |
Cytosine (C)
|
EXTRA CREDIT PCR IMAGE
(CREDIT: http://agctsequencing.wordpress.com/tag/pcr/)
|