BME100 s2014:T Group16 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||
|
OUR TEAM
LAB 1 WRITE-UPInitial Machine Testing
Experimenting With the Connections When we unplugged the LSD Screen(part 3) from the motherboard(part 6), there was no information displayed on the screen. When we unplugged the white wire that connects the motherboard (part 6) to heat block(part 2)which allows for correct temperature measurements, the machine was unable to record the accurate temperature. Test Run The initial test run was conducted on March 20, 2014. After downloading the software, we observed the internal and external features of the PCR machine. We conducted a test run in which we used empty tubes and set the temperatures as prescribed in the lab manual. The initial run was completely unsuccessful. The machine had only heated up to a temperature of 62 degrees and no cycles were initiated. After about 10 minutes, we stopped the experiment due to suspicion of smoke and smell. Upon opening, we found that the tubes had melted. The experiment is marked as a failure as the machine had failed to complete any cycles or reach the desired temperatures, causing the tubes to overheat and melt. One possible error worth noting, was the possibility of the top screw being over-tightened adding pressure on the inner plate and resulting in the melting of the test tubes.
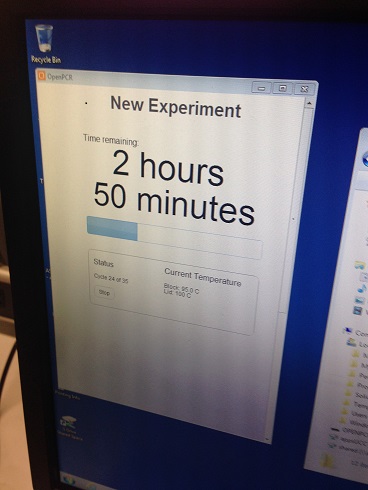
ProtocolsThermal Cycler Program The program was set as follows: -HEATED LID: 100°C -INITIAL STEP: 95°C for 2 minutes -NUMBER OF CYCLES: 35 -Denature at 95°C for 30 seconds -Anneal at 57°C for 30 seconds -Extend at 72°C for 30 seconds -FINAL STEP: 72°C for 2 minutes -FINAL HOLD: 4°C
DNA Sample Set-up
1. Preparing required materials that are 8 empty PCR tubes ,PCR reaction mix ,template DNA, positive and negative control and 200 μL micropipettor 2. Labeling the 8 tubes and placing them in two different rows ( four in each row) 3. transferring 50 μL of the given PCR reaction mix into the eight labeled tubes by using the micropipette 4. Adding the negative and the positive controls to the first two of the tubes which are place in different rows. 5. Transferring DNA of patient 1 to the next three tubes(A2,A3,A4) of the first row which its first tube contains positive control , and DNA of patient 2 to the next three tubes (B2,B3,B4) of second row which its first tube contains negative control 6. Close the lids tightly on your PCR reaction tubes 7. Placing tubes in PCR machine
DNA/ primer mix Patient’s DNA Forward primer and reverse primer
Research and DevelopmentPCR - The Underlying Technology Collecting data for Pipette Practicing: We observed both the final color and volume in micro-liters and recorded that as data. The color observations are our qualitative data and the measurements of the volumes are our quantitative data. In engineering, accuracy is defined as the closeness of measurements of a quantity to that of the actual value (or expected value or outcome) and precision describes the consistency of the results, regardless of its accuracy. Accuracy can happen, also, regardless of precision, as we may achieve the desired data, but not achieve it consistently. This case is true for our experiment with the pipette practicing, as our results were inconsistent.
(BONUS points: Use a program like Powerpoint, Word, Illustrator, Microsoft Paint, etc. to illustrate how primers bind to the cancer DNA template, and how Taq polymerases amplify the DNA. Screen-captures from the PCR video/ tutorial might be useful. Be sure to credit the sources if you borrow images.)
| ||||||||||||||