BME100 f2017:Group9 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
Heated Lid: 100 CHEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25 Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
Research and DevelopmentPCR - The Underlying Technology
The base-pairing occurs during the anneal and extend steps of the cycle. The primer attaches to a complementary base during the anneal. The extension step pairs all the bases of the DNA strand starting from the primer.
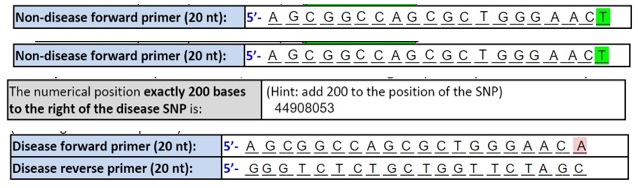
SNP Information & Primer DesignBackground: About the Disease SNP SNP stands for single nucleotide polymorphism. It accounts for the large portion of genetic variation between individuals by differing the nucleotides in certain sections of the DNA. Most of the time, SNPs are used as biological markers and do not have a damaging effect on the individual. However, when the SNPs lie close to or within a gene they are more likely to play a direct role in disease. The specific SNP, rs769452, is found in Homo sapiens on the chromosome 19:44907853. The clinical significance of this SNP is that it is pathogenic and is linked to Alzheimer's Disease. APOE stands for Apolipoprotein E and is the gene where the SNP rs769452 is located. APOE is responsible for carrying cholesterol through the bloodstream. Three unique terms include amyloid-beta binding, antioxidant activity, and cholesterol binding. An allele is the specific location of a gene on a chromosome. The non-disease allele has the codon CTG while the disease allele has the codon CAG. The numerical position of the SNP is 44907853. Primer Design and Testing The results of the primer test showed that the sequence of DNA that would be replicated would be what lies in the 5' to 3' direction after the forward primer and would end before the start of the reverse primer. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||