BME100 f2017:Group8 W0800 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
|
OUR COMPANY
PCR People LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System BME 100 split into 17 teams to diagnose 34 patients (2 patients per group). To find a certain DNA strand of the specific disease, a PCR machine along with necessary nucleotides and disease primers were used to undergo several DNA replication processes with repeated heating and cooling to amplify the disease-ridden strand. With this process, the non-disease DNA will be relatively small in amount. Then a fluorimeter and the software ImageJ were used to analyze the the solution samples. After analyzing the solutions, the data from all of the groups with the respective patients were gathered for analysis using Bayesian statistics. To prevent error in each steps, the PCR machine ran numerous cycles of heating and cooling, allowing for numerous DNA replications with the disease-ridden primer and the disease along with the nucleotides. In ImageJ calibration, all of the samples had pictures taken with the same setup with a couple of trials. In this BME 100 class, the 20 patients concluded positive, 48 patients concluded negative, and 2 patients came out to be inconclusive. There was a group that did not have any data for its respective patients, but the lack of data was not included. Challenges for the PCR lab in human errors such as not measuring the amount of solutions properly with the micropipettors and not keeping the set-up for the pictures correctly. Some calculation errors include not using the exact value. Although these errors may have popped up, due to the use of PCR machine running many runs, and with multiple trials, these errors were minimized. What Bayes Statistics Imply about This Diagnostic Approach
One source of error that could have affected the outcome of the PCR process is that when using the flourimeter, the phone could have tilted or fell, and this would have led to inaccurate measurements. Another sources of error could have been during the flourimeter process a group switch the phone that they were using because this would have led to different results because each phone would have different settings and camera capabilities. Another source of error during the PCR process could have been when using the micropipetter, not all of the solution could have been released into the plastic vials. This would have led to less DNA being replicated and would have changed the final results for the reaction. Intro to Computer-Aided Design3D Modeling Our Design
Feature 1: ConsumablesThe prepackaged kit will be easy for the user as it will have the most important items for the experiment. The kit will be made specifically for the PCR machine. The heating block for the PCR machine has been altered making the plastic tubes a different size. To make this easy for the user the kit will have the tubes that specifically fit the PCR machine. The DNA for the patient will already be in the plastic tubes to reduce waste. The other solutions (PCR mix, primer solution, SYBR Green Soltuion, buffer soultion) will be included in the package in their specific vial pre-labeled. The pre-labeling will make the experiment run easier for the user and help reduce human error for proper testing. The SYBR Green Solution will be wrapped in tin foil to eliminate the solution's exposure to the sun. The micropipettor will be included as well since this consumable is going to be used the most. The tips for the micropipettor will be placed in a smaller soft plastic box than the one used in class to reduce space within the kit. There will be enough tips for the experiment and a couple extra just in case. This is all that is needed for the PCR machine to run the machine to multiply the certain section of the DNA. There will also be the glass slides that will be used during the fluorimeter use, there will be 5 slides which can be used for 25 tests. Example Photos: Pre-labeled vials/plastic tubes Micropipette tips in soft plastic box for easy disposal https://www.universalmedicalinc.com/brand-non-sterile-ultra-micro-pipette-tip-0-5-20ul.html
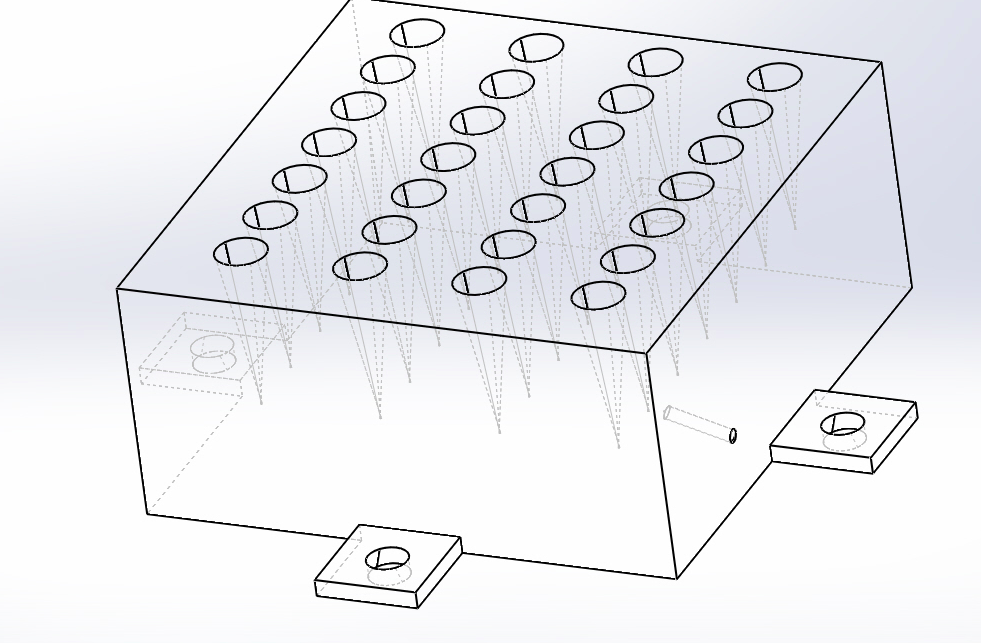
Feature 2: Hardware - PCR Machine & FluorimeterIn the new system, the PCR machine was updated while the fluorimeter remained the same. Compared to the old design of the PCR machine that could fit up to sixteen plastic vials, the updated design can now fit up to 28 plastic vials. With this change, the height of the PCR machine was increased, allowing for the depth of the vial slots to increase from 11 mm to 17 mm. Although this does not account for the 1.25 mm decrease in diameter of the vial slots, it is possible to fit more plastic vials within the machine, thus processing more samples within the same amount of time. The design of the fluorimeter will remain the same, and within the system, will be used the same as paired with the PCR machine.
| ||||||