BME100 f2017:Group8 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
(http://www.promega.com/resources/protocols/product-information-sheets/g/gotaq-colorless -master-mix-m714-protocol/)
have the same forward primer and reverse primer
the samples will become cross-contaminated
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes (The DNA will start to unwind) NUMBER OF CYCLES: 25 DENATURE at 95°C for 30 seconds, Anneal at 57°C for 30 seconds (Now the DNA will be into two strands) EXTEND at 72°C for 30 seconds (Now the DNA polymerase will bond to the DNA and start pairing nucleotides) FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
Research and DevelopmentPCR - The Underlying Technology Q1. What is the function of each component of a PCR reaction?During the PCR process, template DNA will function as a base for the replication process to occur. Once the template DNA is set then the primers will attach to specific sites on the template DNA, these primers will give the binding location for DNA polymerase. The DNA polymerase will bind to the primer and then will utilize the nucleotides present to complete the other portion of DNA after the DNA was split. Question 2 What happens to the components (listed above) during each step of thermal cycling?INITIAL STEP: 95°C for 2 minutes:Heating the samples to 95 degrees Celsius makes the DNA's double helix begin to unwind. Denature at 95°C for 30 seconds:The DNA now is becoming twice as many single-strands than the initial sample amount. This is known as denaturing. Anneal at 57°C for 30 seconds:Now that the sample is cooled to 57 degrees Celsius, the primers have already attached to their target sequence, and made the single strand a double-helix. This leaves the original DNA to remain as single-strands, and the DNA samples separate again, leaving the sample with twice as many target DNA strands. Extend at 72°C for 30 seconds:At 72 degrees Celsius, the polymerase will attach itself to the target DNA and begin pairing bases until it reaches the primers on the end of the strands. FINAL STEP: 72°C for 2 minutes:This is to make sure the single-stranded DNA is completely extended, and the completed replications of the initial DNA sample are in the form of a double-helix. FINAL HOLD: 4°C:This temperature of 4-degree Celsius is used to store the sample for a short period of time.
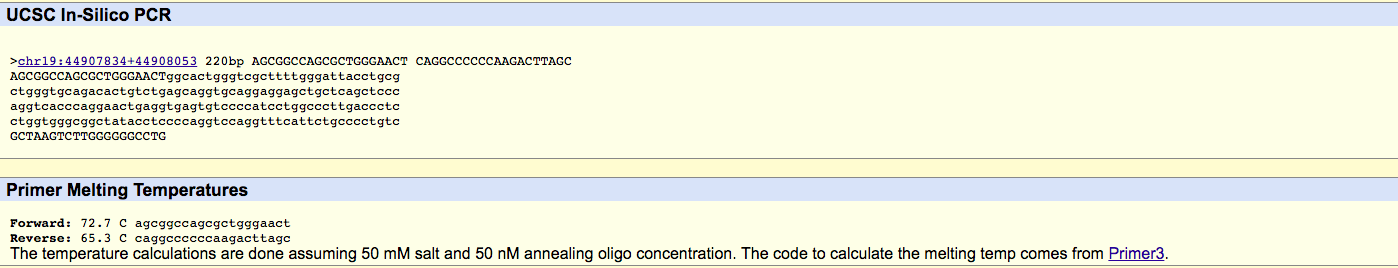
Q3. Nucleotide Base-pairingAdenine(A)-Thymine(T) Thymine(T)-Adenine(A) Cytosine(C)-Guanine(G) Guanine(G)-Cytosine(C) Q4. During which two steps of thermal cycling does base-pairing occur?The steps in which the temperature of the solution is increased to 72 degrees Celsius and base-pairing can occur by the DNA polymerase in a process called elongation. At the end of the cycles, final elongation will take place to make use of all nucleotide base-pairs. SNP Information & Primer DesignBackground: About the Disease SNP SNP are single nucleotide polymorphisms which are a type of genetic variation between people, essentially SNPs are nucleotides. Each person has approximately 3 million SNPs but out of the millions, SNPs only occurs in approximately 1 out of every 300 nucleotides. What makes SNPs special is that they can be used as genetic markers, which help identify genes that are associated with a disease. SNPs can be used to predict a person susceptibility to addiction, etiology of diseases, and determine hereditary diseases within a family. On the other hand, if an SNP occurs in a gene it may actually be a factor for which a disease is developed which in turn begins to affect the gene's functionality. The identification of the diseases can be found through a Genome-Wide Association Study (GWAS) which identifies recurrent SNPs in a persons genetic sequence. Primer Design and TestingWhat is a nucleotide? A nucleotide is the building block for DNA. It is the smallest piece of DNA. It varies from A, T,C, and G. What is a polymorphism? Polymorphism in DNA is the difference in the sequence of DNA among individuals. What species is this variation found in? Homo Sapiens What chromosome is the variation located on? 19:44907853 What is listed as the Clinical significance of this SNP? Pathogenic What condition is linked to this SNP? Alzheimers What does APOE stand for? Apolipoprotein E What is the function of APOE? This gene will provide instructions that will lead to the creation of Apolipoprotein E Write the first three unique terms you see. Amyloid-beta binding, protein binding, phospholipid binding. What is an allele? An allele is a gene located in a specific area along a chromosome. They determine a trait in an individual. The disease-associated allele contains what codon? CCG The numerical position of the SNP is: 44907853 Non-diseaseFor this part of the lab we designed a non-disease forward primer at the position of 44907853. Then we designed a non-disease reverse primer by adding 200 to the original position to simulate two primers amplifying the DNA. Non-disease forward primer 5’AGCGGCCAGCGCTGGGAACT3’ Non-disease reverse primer position 44908053 5’CAGGCCCCCCAAGACTTAGC3’ Results Non-disease
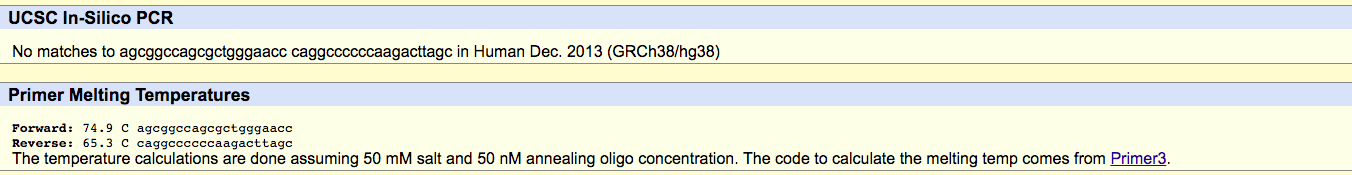
DiseaseFor this part of the lab we designed a disease forward and reverse primer by simply taking the designed non-disease forward and reverse primers and changing the last base on the forward primer to the disease. Disease Forward Primer 5’AGCGGCCAGCGCTGGGAACC3’ Disease Reverse Primer 5’CAGGCCCCCCAAGACTTAGC3’ Results Disease
| |||||||||||||||||||||||||||||||||