BME100 f2017:Group7 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
DNA Sample Set-up Procedure
OpenPCR program
Research and DevelopmentPCR - The Underlying TechnologyFunctions of Components in a PCR ReactionThere are four components in a PCR reaction, which include template DNA, primers, Taq Polymerase, and Deoxyribonucleotides (dNTP’s). The function of template DNA is to be the subject sample that is analyzed for the experiment and later replicates. The function of primers 1 and 2 is to attach at the opposite ends of split DNA fragments and provide a location for DNA polymerase to later bind. The function of Taq Polymerase is to withstand high temperatures, bind to the primer sites, and collect free floating nucleotides to attach to the DNA until the strand is complete with base pairs. The function of Deoxyribonucleotides (dNTP’s) is to create DNA copies through base pairing from the Taq Polymerase. PCR Steps    Base PairingBase pairing occurs during the steps to anneal at 57°C for 30 seconds and extend at 72°C for 30 seconds. During annealing, primer attaches to a DNA strand through base-pairing. During extending at 72°C, DNA polymerase becomes activated and grabs nucleotides from the PCR mixture to attach at a strand specified by the primer. Bases that anneal to each other are as follows, Adenine (A) to Thymine (T) and Guanine (G) to Cytosine (C).
SNP Information & Primer DesignBackground: About the Disease SNPIn this part of the lab, the NCBI database was used to find a disease-associated sequence. Key terms related to this search are nucleotides and polymorphisms, as the database enables researchers to locate short genetic variations. Nucleotides are the backbone of nucleic acids, consisting of phosphate connected to sugar connected to a base pair. A polymorphism is a genetic variation found in DNA that usually will divide the population of a species into two or more different phenotypes of the species. The specific single nucleotide polymorphism investigated in this lab was a variation found in chromosome 19:44907853 of the Homo sapiens species. The clinical significance of this SNP is that it is pathogenic and is associated with both Alzheimer’s Disease and stroke. To analyze this SNP, the DNA sequence of the SNP and the surrounding sequence were assessed. The gene corresponding to this SNP is apolipoprotein E (APOE), which has many functions including amyloid-beta binding, antioxidant activity, and cholesterol binding. The disease-associated allele of this gene is CCG. An allele is the different form a gene can have at the same place on a chromosome. It is a variation that occurs through mutations. Primer Design and TestingFor the primer test, a non-disease forward primer was first designed for rs769452 based on the numerical position of the SNP (44907853). The forward primer is 5’-AGCGGCCAGCGCTGGGAACT-3’. To design the reverse primer, the reverse strand of the sequence was recorded 200 bases to the right of the disease (44908053). The reverse non-disease primer is 5’-CAGGCCCCCCAAGACTTAGC-3’. Disease SNP-specific forward and reverse primers were designed by substituting the final base of non-disease forward primer. This makes it the same as the disease SNP nucleotide. The disease reverse primer is 5’-AGCGGCCAGCGCTGGGAACC-3’ and the disease reverse primer is 5’-CAGGCCCCCCAAGACTTAGC-3’. The primers were tested by inputting them into the UCSC In-Silico PCR website. The non-disease primers search result showed a 220 bp sequence from the chromosome 19:44907853 (chr19:44907834 through 44908053), which signaled that the primers do in fact work. The submission with disease-specific primers resulted in no match, which signaled that the disease-specific primers were correctly designed. There was no match because the changed allele doesn’t occur in a non-diseased genome, which is what this database uses as a reference. 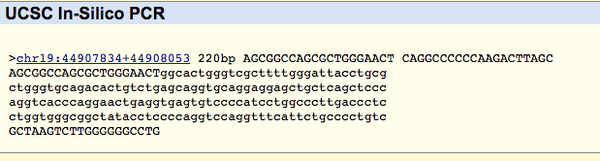  |
|||||||||||||||||||||||||||||||||




