BME100 f2017:Group7 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
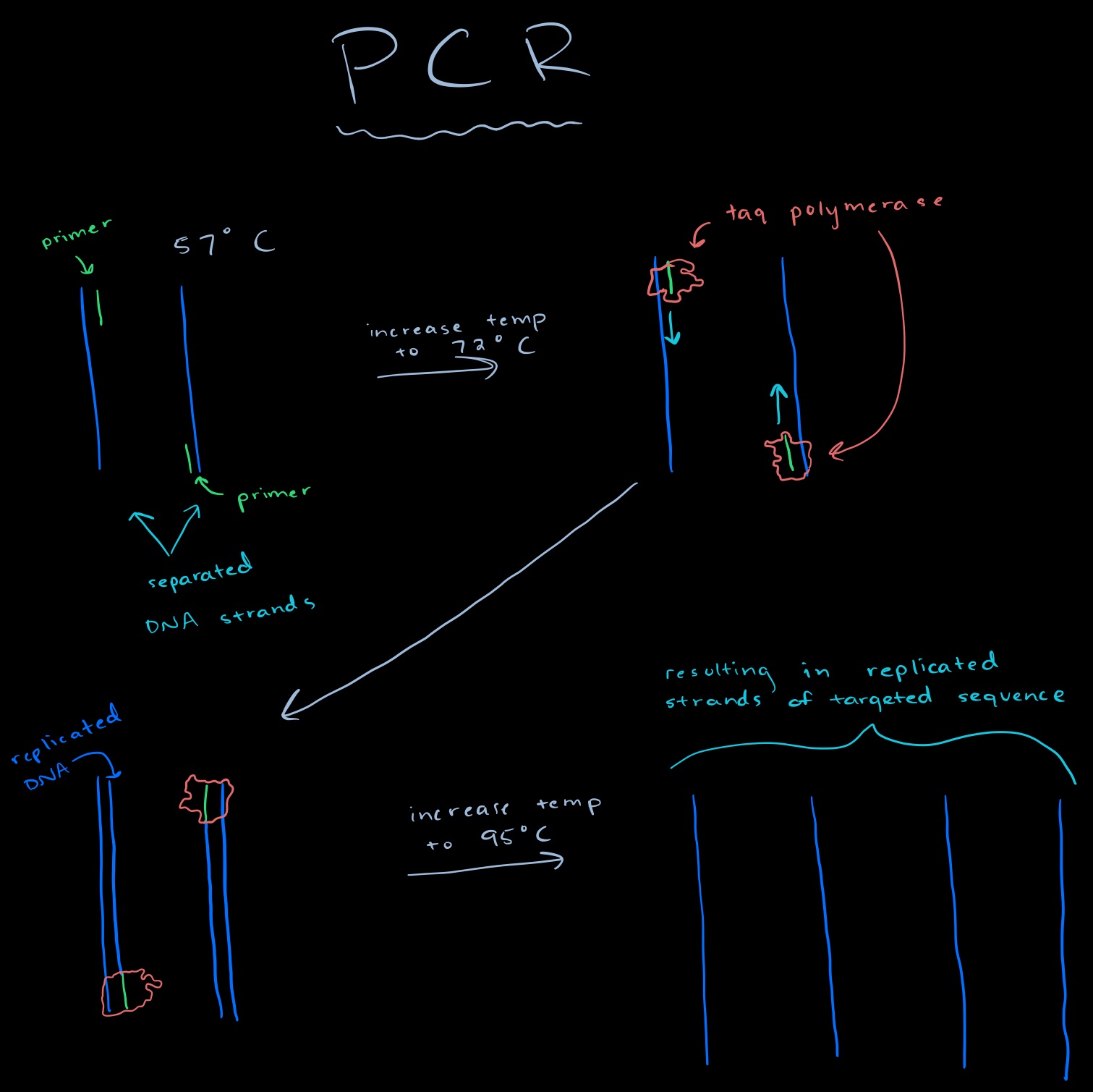
The PCR program is run by having the heated lid at 100 degrees Celsius. The initial step is set the program to 95 degrees C for 2 minutes. This will be done for 25 cycles, denaturing at 95 degrees C for 30 seconds, annealing the DNA at 57 degrees C for 30 seconds, and extending the DNA at 72 degrees C for 30 seconds. The final step is the keep the sample at 72 degrees C for 2 minutes, and set the final hold to 4 degrees C.
Research and DevelopmentPCR - The Underlying Technology Function of Each Component The DNA sample acts as a template for replication. The goal of PCR reactions is to target a certain sequence of DNA and replicate it; This sequence is located on the template DNA. Primers are short RNA sequences specific for the targeted DNA sequence. PCR utilizes 2 primers: reverse primer and forward primer. Primers are designed in a way that they contain the paired sequence of the beginning and end part of the desired target sequence and can bind to the DNA sample. Taq polymerase is a DNA polymerase obtained from heat-resistant organisms. The role of DNA polymerase is to attach to both primers and elongate the targeted sequence of DNA. For this process to happen, dNTP's need to be added to the mixture for DNA polymerase elongation. Steps of Thermal Cycling During the first step of this procedure, the sample is slowly heated to 95 C. This process weakens the intermolecular forces between base pairs located between 2 DNA strands. In the denaturing step, hydrogen bonds and other intermolecular forces break and 2 strands of the template DNA are separated. During the annealing phase, the sample is cooled down to 57 C for 30 seconds. This step enables primers to bond to the targeted DNA sequence. Primers are designed to contain the complementary sequence of the beginning and ending target sequence of DNA. During the annealing step, primers locate their complimentary sequence on the DNA (beginning and end of the targeted sequence at the 5' and 3' end) and form hydrogen bonds with the complimentary strand through base pairing. Base pairing occures between A -T and G-C. At the extension step, the mixture is reheated to 72 C for 30 seconds. During this step, DNA polymerase attaches to the previously added primers and creates a new strand of DNA. DNA polymerase cannot initiate DNA synthesis without an initial primer and will only add nucleotides to the 3' end of primer. Once attached to the primer, DNA polymerase uses the dNTP's in the mixture and pairs them with the complimentary nucleotide on the DNA. In other words, it pairs each T with an A and each G with a C. After the process is finished, DNA polymerase is removed from the newly synthesized DNA strands, the mixture is cooled down to 4 C and new hydrogen bonds are formed between DNA strands thus creating a newer, shorter DNA. As mentioned previously, Base pairing occurs a the annealing and extension phase of PCR. During the annealing phase, nucleotides on the primer pair with corresponding sequences on the DNA thus making a hybrid structure. In the extension phase, DNA polymerase uses dNTP's from the sample to make a complimentary strand for each template strand of DNA. SNP Information & Primer DesignBackground: About the Disease SNP The basic building blocks of DNA are nucleotides (Adenine, Guanine, Thymine, Cytosine). When a mutation in one of these pairs along the DNA molecule occurs, it is called a single nucleotide polymorphism (SNPs). Some of these SNPs can cause severe damage to the function of the cell if it occurs in an area of an important gene. The disease SNP we are looking at today is due to a nucleic acid change of a Thymine to a Cytosine in chromosome 19, location 44907853. It is pathogenic meaning it is caused by either viruses, bacteria, or other types of germs and only affects homo sapiens. This SNP is located on a gene that codes for Apolipoprotein E (APOE) which is a protein that is essential for the normal catabolism of triglyceride-rich lipoprotein constituents. Specifically, it is responsible for Amyloid-beta binding, antioxidant activity, and cholesterol binding. Studies have shown that this allele of the APOE gene is linked to conditions of Alzheimer’s and stroke.
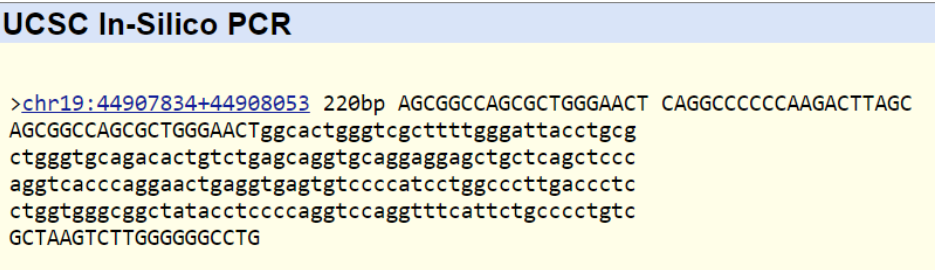
Each PCR reaction needs both a forward primer and a reverse primer. The forward primer has the SNP as the 20th and last base in its sequence and the reverse primer begins 200 bases right of the SNP. Both of these primers need to be read from the 5' to 3' direction. The following primers are from the healthy and mutated nucleotide sequence of the disease mentioned above Forward Non-disease Primer: 5’- A G C G G C C A G C G C T G G G A A C T - 3’ Reverse Non-disease Primer: 5’- C A G G C C C C C C A A G A C T T A G C - 3’ Forward Diseased Primer: 5’- A G C G G C C A G C G C T G G G A A C C - 3’ Reverse Diseased Primer: 5’ - C A G G C C C C C C A A G A C T T A G C - 3’
The non-diseased primer matched with a sequence of DNA. As for the diseased primer, there was no DNA sequence that matched it. This was the expected result of the test as a diseased primer would have no match with a healthy human genome.
| ||||||||||||||||||||||||||||||||||