BME100 f2017:Group6 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
OpenPCR program
Research and DevelopmentPCR - The Underlying Technology
Components of PCR When performing a PCR, a Template DNA is needed. The DNA template contains the targeted DNA that is to be sequenced and can be found in saliva, blood, or a piece of hair, it only requires a very small amount1. During the reaction, primers locate the target DNA by attaching one primer to both ends of the targeted DNA1. DNA Polymerase is added to grab nucleotides from its surroundings and attach them to the proper sites to create DNA copies1. In PCR though, taq polymerase is used because it is found in an organism that can whithstand high temperatures, which is useful since PCR undergoes extremely hot temperatures1. Deoxyribonucleotides are the A’s C’s T’s G’s that make up the genetic code1. Steps of PCR PCR undergoes three major steps, denaturation, annealing and extension. In the first step, the temperature is set to 95 degrees celsius, the extreme heat causes the DNA to denature and separating the double helicase into two single stranded DNA1. In annealing, the temperature cools to 50 degrees celsius, the primers then come in and attach to the ends targeted DNA1. The third step, the temperature is set to 70 degrees celsius, the DNA polymerase comes in and locates the primers1. It then starts attaching complementary nucleotides, A codes to T and T codes to A and C codes to G and G codes to C, and when it gets to the end of the targeted DNA it falls off1. The DNA is stored and cooled in a 4 degree celsius environment. Steps two and three are repeated, where base pairing occurs, until millions of copies of the targeted DNA are produced1. 1. http://learn.genetics.utah.edu/content/labs/pcr/
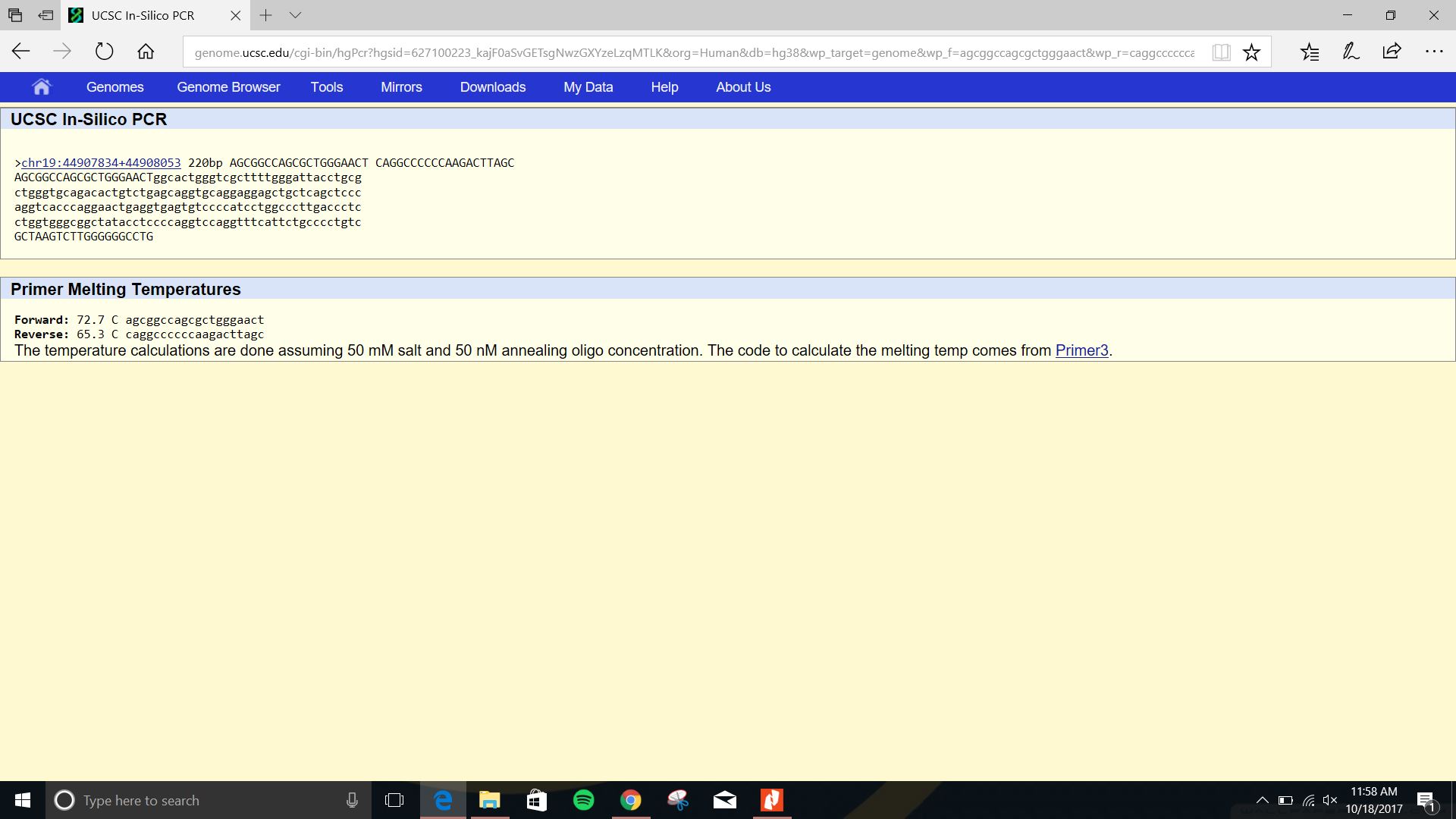
SNP Information & Primer DesignBackground: About the Disease SNP A single nucleotide polymorphism also known as a SNP is the most common form of genetic mutation. It occurs when a single nucleotide is changed. When these SNP’s are the cause of a disease they are called disease SNP’s. Disease SNP’s most commonly occur in the DNA between genes and can act as markers for a disease. This is helpful because it more easily allows researchers to determine where a disease is in DNA. Primer Design and Testing The Results of our summary test came back positive. This means that we correctly identified the location of the desired gene and we correctly identified the disease SNP.
| |||||||||||||||||||||||||||||||