BME100 f2017:Group5 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25 Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
Research and DevelopmentPCR - The Underlying Technology There are 4 main components of a PCR reaction: template DNA, primers, Taq polymerase, and dNTPs. The template DNA is the dsDNA you are that contains the sequence you are intending to amplify. Primers are short strands of DNA made in the lab. They can have any sequence of nucleotides necessary to complement the regions flanking the part of the template DNA you are interested in amplifying. Taq polymerase is the enzyme that adds nucleotides one at a time to the 3' OH groups of a DNA strand, complementary to the template strand. dNTPs are the building blocks of DNA that are used to extend DNA strands in PCR. PCR begins with an initial denaturation at 95 degrees C for 30 seconds, to begin to denature the dsDNA into 2 strands. Then a cycle of denaturation, annealing, and extension repeats for the desired number of cycles to achieve optimal amplification. This cycle includes denaturation at 95°C for 30 seconds, which separates dsDNA strands, annealing at 57°C for 30 seconds, during which primers anneal to dsDNA, and extension at 72°C for 30 seconds, during which Taq polymerase extends the primers using available dNTPs in solution. Finally, after the desired number of cycles, the reaction is held at 72°C for 2 minutes, to ensure that the ssDNA is fully extended by Taq polymerase (this is the optimal temperature for Taq pol to work). Finally, the products can be stored in the short-term at 4°C. Base pairing thus occurs during the annealing and extension steps. During annealing, the primers anneal to ssDNA through complementary base pairing. During extension, dNTPs from solution are base paired by Taq polymerase to the template strand, extending off of the 3' OH group from the strand begun by the primer. A pairs to T and G pairs to C.
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is a basic building-block of DNA, made up of a nucleotide attached to a phosphate group. A polymorphism is a genetic variation, such as a differing nucleotide between two individuals. We spent this lab investigating the SNP rs769452, which is found in Homo sapiens, and located on chromosome 19 (19:44907853). This SNP is pathogenic, and is associated with Alzheimer's disease. It is a SNP on the apolipoprotein E (APOE) gene. APOE codes for a protein that is amyloid-beta binding and has antioxidant activity. An allele is a specific form of a gene found at a certain location on a chromosome, of which there are multiple, distinguished by mutations. The disease-associated allele contains the codon CCG, while the non-disease allele contains the codon CTG.
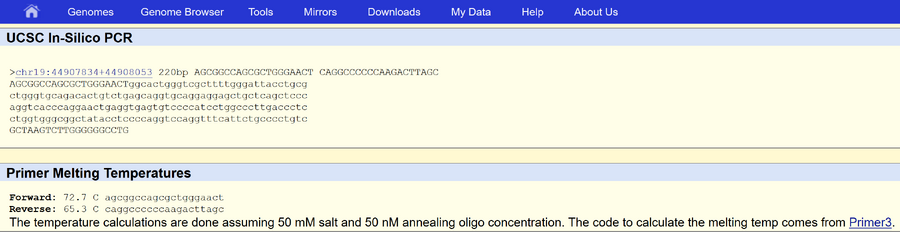
The numerical position of the SNP is 44907853. Our non-disease forward primer, which we made from the sequence 20 bases long that ends with the nucleotide at the position of our SNP, is 5’ AGCGGCCAGCGCTGGGAACT 3’. Our non-disease reverse primer begins at position 44908053: 5’ CAGGCCCCCCAAGACTTAGC 3’
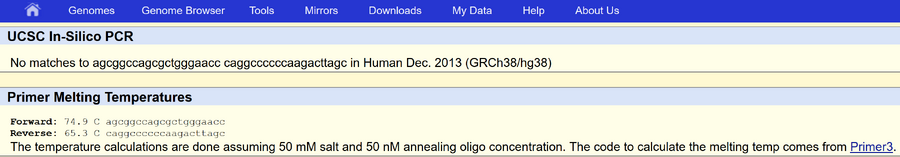
Our disease-specific primers are as follows. Forward: 5’ AGCGGCCAGCGCTGGGAACC3’ Reverse: 5’ CAGGCCCCCCAAGACTTAGC 3’ | |||||||||||||||||||||||||||||||||