BME100 f2017:Group1 W1030 L6
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||
|
OUR COMPANY
Our Brand Name LAB 6 WRITE-UPBayesian StatisticsOverview of the Original Diagnosis System BME 100 tested patients for the disease-associated SNP by running various PCR experiments and analyzing the photos of DNA samples with amplified replicants of data for various patients. This process was split among sixteen groups of students, with each group testing two patients (three DNA replicates per patient). The approach can be summarized as follows: within each group, two patients were assigned and three replicates of each patient’s DNA was tested. To complete the PCR reactions for both patients, the following materials were added together in a test tube which was then inserted into a thermocycler for completion of the experiment: replicate DNA (specific to each patient), PCR reaction mix and DNA primer mix. After placement in the thermocycler, the PCR reaction was completed, where DNA replicates were replicated billions of times. There were a total of six PCR reactions for every group (for 16 groups) and a total of 32 patients. To ensure that error was minimized during these experiments, the exact micropipetting technique was used, and correct measures were followed to ensure proper measurements made within each test tube and proper disposal of all materials. Additionally, during the ImageJ analysis portion of these experiments, pictures were taken using the proper camera settings (as designated by the lab workbook) and through the use of a fluorimeter, where the flap was closed so each DNA sample- paired with SYBR Green 1 for mutation identification- was seen as clearly as possible. Three pictures were taken of both 1) the six calibration concentrations (seen in lab write-up 5) and 2) both positive/ negative controls and all six replicate DNA samples for both patients. These photos were analyzed in ImageJ to determine RAWINTDEN values and therefore, both quantitatively and qualitatively, the results of disease presence could be determined with as little error as possible. Three replicate DNA samples were used for each patient, and three pictures were taken for each sample, to minimize error within the experiment.
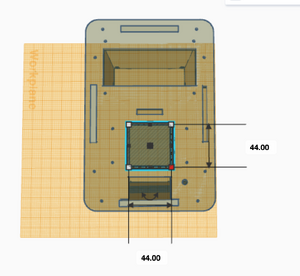
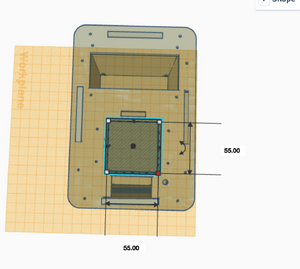
What Bayes Statistics Imply about This Diagnostic Approach Calculations 1 and 2 Calculations 1 and 2 refer to the sensitivity and specificity of the system (individual PCR replicates) respectively, with regard to the ability of detecting the disease SNP. Calculation 1 reveals a very high probability (close to 100%) that a patient will get a positive final test conclusion, given a positive PCR reaction. Calculation 2 reveals an even higher (very close to 100%) probability that a patient will get a negative final test conclusion, given a negative diagnostic signal. Therefore, the individual PCR replicates are very reliable in terms of detecting the disease SNP. Calculations 3 and 4 Calculations 3 and 4 refer to the sensitivity and specificity of the system (individual PCR replicates) respectively, with regard to the ability of predicting the disease. Calculation 3 reveals a moderate probability (close to 50%) that a patient will develop the disease, given a positive final test conclusion. Calculation 4 reveals an extremely high (100%) probability that a patient will not develop the disease, given a negative final test conclusion. Therefore, individual PCR replicates are more reliable in diagnosing when the diagnosis is negative, rather than positive. About half the time, a positive diagnosis is effective in predicting the development of the disease. Sources of Error Sources of error lie in inaccurate pipetting prior to the PCR reaction process, where inaccurate volumes of PCR reaction mix, DNA primer mix or the replicates themselves could have been added to the test tubes, or the incorrect mix could have been added (human error). Additionally, error may have occurred during the thermocycler process, or in the imaging process where the fluorimeter may have been partially open, allowing light to pass through the reaction mix and the SYBR Green 1 when taking photos to detect the mutation. These errors may have led to false detection of the presence of the mutation- the disease SNP- in each replicate and thus, Bayes values may have been influenced. Intro to Computer-Aided Design3D Modeling Our Design Our new design does not change much of the initial design that was provided to us. We saw very little that could be done to vastly improve the machine in a practical way. Our group ended up deciding that one of the biggest downfalls to the initial design was that you could only run 16 PCRs at once. To combat this, we decided to increase the capacity size of the tube holder from a 4x4 box to a larger 5x5 box. This would allow for 25 PCRs to be run at a time instead of a measly 16. This increase in size would not affect heating and cooling, as the temperature is presumably controlled by an electric heating element, which is accurate even over a large area. Also, the design with the bigger area for the PCR tubes does not effect any other of the components or holes in the design, making it a small but ideal change that could boost the productivity of a single Open PCR by 56%.
Feature 1: ConsumablesBecause our new design—a thermocycler with the capability of running 25 reactions at a time (rather than the original 16)—has more open space and requires more tubes for a single reaction, our consumables kit will package 25 tubes and more pipette tips. Additionally, the volume of PCR mix and primers included in the consumables kit will be doubled, because each kit must have the materials necessary to run more reactions at one time. The number of tips will not be doubled from its original size, however, because there were too many tips in the consumables kit from BME 100, creating unnecessary costs and waste. The following is a complete list of materials included in the packaged consumables kit for our design:
Feature 2: Hardware - PCR Machine & FluorimeterBoth the Open PCR machine and fluorimeter will be included in our new design, called PCR4U; the PCR machine will have increased capability of running more reactions at one time and the fluorimeter will be designed with a new cradle for a smartphone and a new appendage that is to be folded over the phone during the imaging process. The previous fluorimeter had a closing flap on the outside to keep its inside dark as images were taken of the resulting PCR reaction mixes; however, the new design will have a cut-out within the flap to be folded over the smartphone, where there will be just enough space for the phone to sit beneath the flap and the inside area will remain dark enough for the images to be taken. From this new design, it will be much more efficient for photos to be taken in a dark space without having to set a timer on the camera or fold the flap over the phone. Additionally, this cut-out will be made adjustable to any camera placed on the cradle beneath it, so that the user has the flexibility to use different cameras other than a smartphone.
| ||||||