BME100 f2017:Group1 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
HEATED LID: 100°C INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25 Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
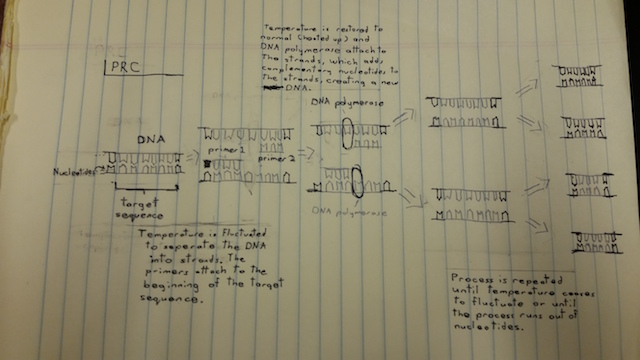
Research and DevelopmentPCR - The Underlying Technology The Function of Each Component: The template DNA is the sample we will amplify and analyze to diagnose the patients. The primers are used to designate which specific sample of the template DNA is amplified. They attach to sites on the template DNA that mark each end of the desired segment. The Taq Polymerase attaches to the primers on each individual strand of DNA and copies the DNA by adding the matching nucleotides to the single strands. It is designed to withstand the high heat of the PCR. The deoxyribonucleotides are used to create copies of the selected segment of the template DNA. Taq Polymerase attaches these to the single, separated strands of DNA in each PCR cycle. What Happens During a PCR Cycle: Initially, the temperature is raised to 95° C for two minutes. Then for 30 seconds, the DNA denatures, meaning the double helix separates into two single-stranded DNA molecules. Then, the temperature cools to 57° C for 30 seconds and the primers lock onto the single DNA strands, preventing the DNA strands from recombining. This is called annealing. Then, the temperature is raised to 72° C over 30 seconds, and the Taq Polymerase is activated. The polymerase adds complementary dNTPs to the strands, starting at the primers and continuing to the end of the strand, where it falls off. This process takes 2 minutes. The cycle will repeat itself for 25 cycles, and then the mixture will be held at 4° C. Base Pairing: When the Taq Polymerase is activated, base pairing occurs. This happens at 72° C in the final stages of the PCR cycle. Adenine and Thymine pair together, and cytosine and guanine pair together.
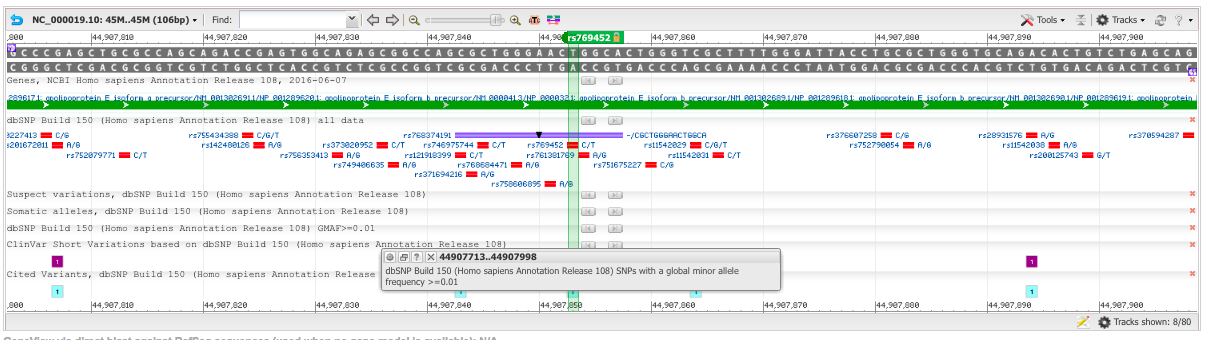
SNP Information & Primer DesignBackground: About the Disease SNP A single-nucleotide polymorphism, SNP, is a variation in a single nucleotide that occurs at a specific position in the genome. At a specific base position in a human genome, a minority of individuals may have a different base than the majority of the population. The two differing nucleotide variations are said to be alleles for this base position. Many diseases, including sickle- cell anemia and cystic fibrosis, are the result of SNPs. Similarly, severity of an illness and one's reaction to treatment can also be dependent on SNPs.
The NCBI database was used to find a sequence of base pairs in the human genome associated with a specific disease. The sequence observed contained a single nucleotide polymorphism (SNP) related to the development of Alzheimer's disease. The numerical position of the specific nucleotide was then recorded so as to be used in designing a primer that would match with the desired sequence. A forward primer was designed based on the location of the nucleotide of interest. Given that every PCR reaction requires two primers to function properly, a reverse primer was needed as well, and designed based on the location of a nucleotide exactly 200 base pairs away from the allele used to create the forward primer. Subsequently, the disease forward primer was designed by changing the final base of the non-disease forward primer to be identical to the disease related SNP. The disease reverse primer was left identical to the non-disease reverse primer. Finally, all four primers were tested in the UCSC In-Silico PCR website to verify proper design function. Non-disease forward primer: AGCGGCCAGCGCTGGGAACT Non-disease reverse primer: CAGGCCCCCCAAGACTTAGC Disease forward primer: AGCGGCCAGCGCTGGGAACC Disease reverse primer: CAGGCCCCCCAAGACTTAGC | ||||||||||||||||||||||||||||||||||