FINAL HOLD: 4°C
Research and Development
PCR - The Underlying Technology
The purpose of the Polymerase Chain Reaction is to amplify a targeted DNA Sequence by making several copies of it. The Polymerase Chain Reaction principally consists of primer mediated enzymatic amplification of the Deoxyribonucleic Acid. It is based on the ability of DNA polymerase to synthesize complementary DNA strands when exposed to a template strand.
The PCR reaction requires the following components in order for it to be carried out:
1. DNA Template: The desired DNA sequence to be amplified
2. DNA Polymerase: The enzyme that facilitates the reaction
3. Oligonucleotide Primers: Short pieces of complementary DNA strands
4. Deoxynucleotide Phosphates: to provide energy for the reaction and are also the building blocks of DNA synthesis
5. Buffer System: To provide optimal condition for the reaction to take place
There are 3 stages to the Polymerase Chain Reaction:
- Denaturation: The first step of the PCR reaction is to heat the DNA strands to 94 degrees Celcius to denature the double-stranded DNA into single-stranded DNA, hydrogen bonds are broken between the strands
- Annealing: The second step of the PCR reaction is to reduce the temperature to 54 degrees Celcius to let Primers to anneal to their complementary sequence in the template DNA
- Elongation: The third step of the PCR reaction occurs at 72 degrees Celcius where the polymerase enzyme sequentially adds bases to the 3′ each primer, extending the DNA sequence in the 5′ to 3′ direction. Thus the new, duplicate strand is elongated
Citation: [1]
“Polymerase Chain Reaction (PCR) : Principle, Procedure, Components, Types and Applications.” LaboratoryInfo.com, Laboratory Info, 28 July 2015, laboratoryinfo.com/polymerase-chain-reaction-pcr/.
Background: About the Disease SNP
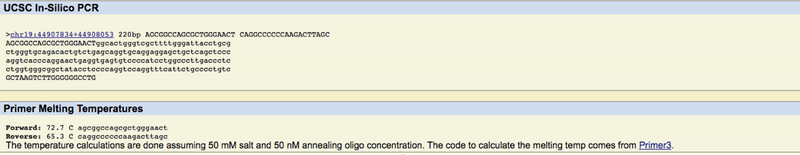
The disease and SNP is found in Homo Sapiens on chromosome 19:44907853. The clinical significance of this SNP is pathogenic and it is linked Alzheimer’s. APOE stands for apolipoprotein E. It acts as a biomarker in the genes of Alzheimer’s disease and Subarachnoid hemorrhages. Alzheimer’s is often caused by the expression of an alternate gene, the allele. The disease-associated allele, in this case, contains the “T” codon with the numerical position of the SNP being 44907853.
Primer Design and Testing
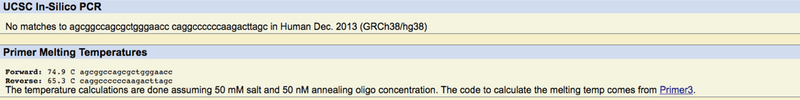
In short, when we wrote our primer codes and ran them through the database, it revealed that the codes we wrote were located on chromosome 19. This is correct because it was revealed earlier that our gene was located on that very chromosome. Additionally, we ran our disease forward primer through the database, it revealed no results, which was the desired result.
Sources
“Polymerase Chain Reaction (PCR) : Principle, Procedure, Components, Types and Applications.” LaboratoryInfo.com, Laboratory Info, 28 July 2015, laboratoryinfo.com/polymerase-chain-reaction-pcr/.






