BME100 f2017:Group12 W0800 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||
|
OUR TEAMLAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
OpenPCR program
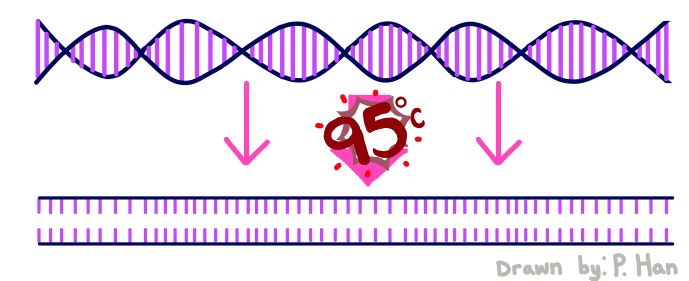
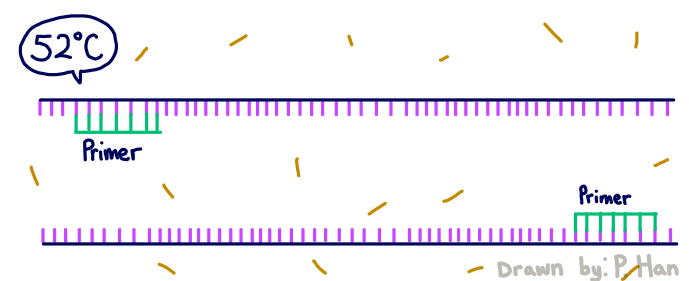
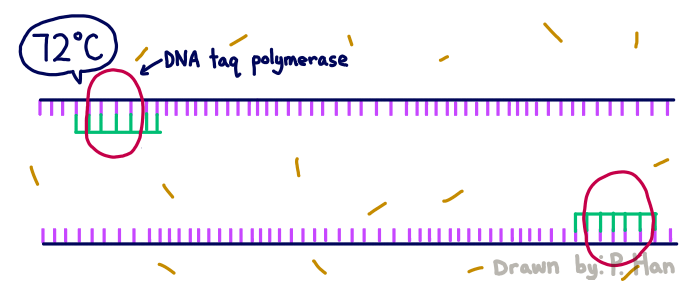
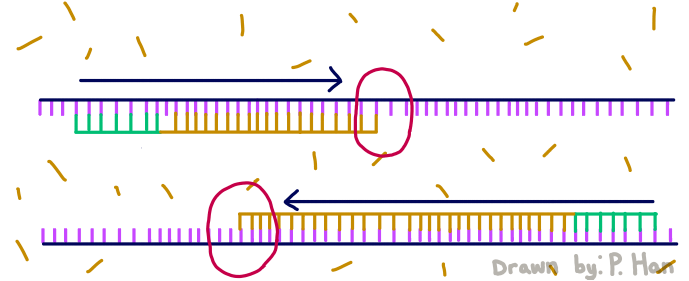
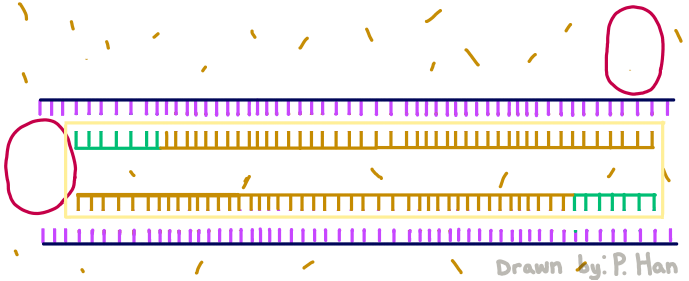
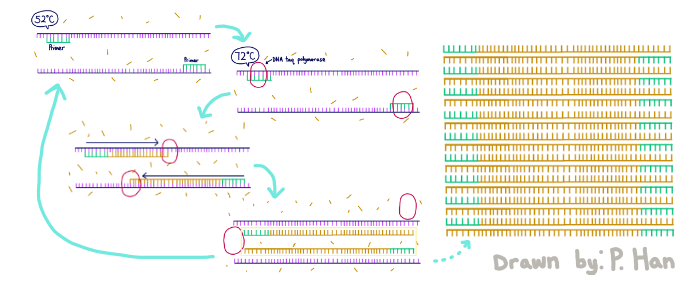
HEATED LID: 100°C Denature at 95°C for 30 seconds, Anneal at 57°C for 30 seconds, and extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes PCR, short for Polymerase Chain Reaction, is a method used by scientists to replicate and amplify the specific region of DNA that is of interest. PCR is done by adding primers to the DNA and repeatedly heating and cooling the mixture which has the effect of simulating the temperatures that are necessary for DNA replication to occur. The first step is for the temperature to go up to 95 degrees Celsius for 30 seconds so that the DNA “denatures,” or splits from its double-stranded form into single strands. After this step, the temperature goes back down to 57 degrees Celsius for 30 seconds which allows the primers to bind to the strands. Then the temperature goes back up slightly to 72 degrees Celsius for 30 seconds so that the enzyme DNA Taq polymerase can attach to the primed sequences and add nucleotides. This is the first cycle and the PCR will happen for 25 cycles in total.
Research and DevelopmentPCR - The Underlying Technology
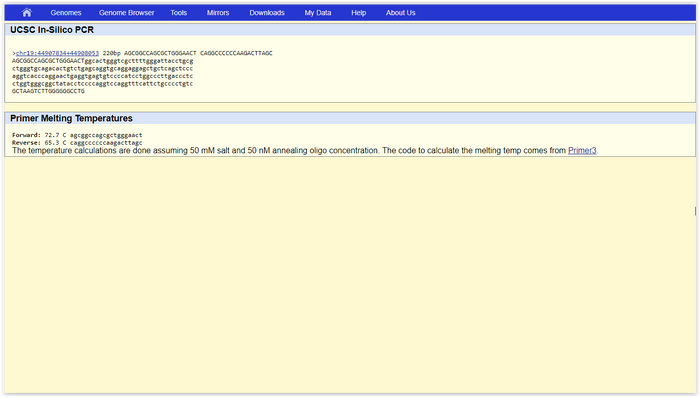
SNP Information & Primer DesignBackground: About the Disease SNP The human genome is made of many long strings of code composed of nucleotides in a specific order, that instruct the cell which proteins to produce. Single nucleotide polymorphisms, or SNPs (which are pronounces like the word "snips"), are a result of an accidental alteration in a single component or nucleotide in the DNA, which can lead to a variation in the proteins produced by the cell. When a piece of DNA is altered in such a way that it causes a change in the proteins produced, that leads to a change in the function of the cell, as proteins are the mechanisms through which the cell works. So when a SNP produces this type of change, it can lead to disease in the cell, and potentially fatal problems in the organism. Due to the fact that these morphisms in DNA can lead to such drastic outcomes in the organism, it is very useful for medical professionals to be able to identify the location of a SNP, so that they have a better understanding of how to approach removal of the disease SNP. Primer Design and Testing (Question 15 on worksheet) In the primer design, we seek to find the forward and reverse primer and read them from 5’ to 3’. We want to find the base pair in the genome that contains the error. This base pair and the 19 base pairs before it form the the forward primer that has a total of 20 base pairs. In this case, the primer started at 44907853 and it was 5’- AGCGGCCAGCGCTGGGAACC. The reverse primer is 200 base pairs prior to the base pair that had the mutation and it serves as a reference point during PCR. Thus, we can find the primer position to be 44908053 by adding 200 from the base pair position of the forward prime and the reverse primer to be 5’- CAGGCCCCCCAAGACTTAGC. We test these primers by inputting them into the genome site and we found the healthy 220-base pair long primer. The results can be found below. When the disease mutation was inputted, we didn’t find the primers since you wouldn’t expect to find it in a healthy human’s genome. | ||||||||||||||||||||||||||||