BME100 f2017:Group10 W1030 L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
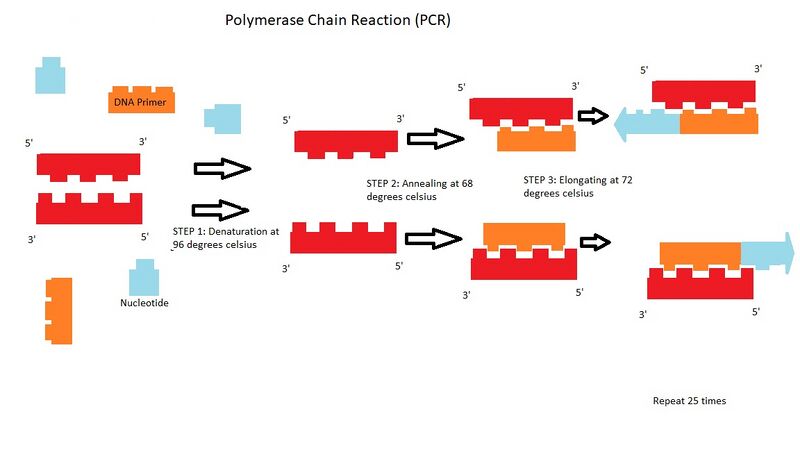
Research and DevelopmentPCR - The Underlying Technology Components of a PCR Reaction In order for a PCR to take place template DNA, primers, Taq polymerase, and Deoxyribonucleotides(dNTP's) must be present. Template DNA provides a template for a new strand to be synthesized. Primers keep the single-stranded DNA from rejoining and provide a marker for Taq polymerase to bind and begin to add nucleotides (extend the DNA).Taq polymerase is an enzyme that adds dNTP's to the end of the strand. Thermal Cycling For the purpose of this description, please see the image above. A DNA double helix (shown in red), DNA primers (shown in orange), and nucleotides (shown in blue) are in solution. In step one of the OpenPCR program, the red double helix separates into two single strands of DNA when the temperature is increased to 96 degrees Celsius; this step is called denaturing. In step two of the polymerase chain reaction, the orange DNA primers attach to the appropriate ends of the single strands of DNA when the temperature is decreased to 68 degrees; this step is called annealing. In step three of the process, the smallest pieces that make up DNA, the blue nucleotides, are added to the ends of the complementary strands of DNA when the temperature is increased to 72 degrees Celsius; this is called elongation. This process is repeated with the two new double helixes formed from the first cycle. Then repeated with all 4 strands created from the first two cycles. Then repeated with all of the strands from the first three cycles and so on.
SNP Information & Primer DesignBackground: About the Disease SNP A nucleotide is a DNA structure that is composed of three parts: a sugar molecule, a phosphate group, and a nitrogenous base. The phosphate group and the sugar molecule covalently bond to form what is know as the “backbone” of a DNA strand. Thymine, cytosine, adenine, guanine and are the four types nitrogenous bases in DNA. Each one typically bonds with one other (thymine to adenine, and guanine to cytosine) through hydrogen bonds which, as a result, form the double helix structure of DNA. While DNA may be comprised of the same four nucleotides, the sequencing of nucleotides are all different, thus creating a wide variety of gene variation or Polymorphism. Polymorphism is a term used in biology, or genetics more specifically, to describe multiple forms of a single gene that exists in an individual or among a group of individuals. For example ear lobes are an example of polymorphism because it separate its species into two groups, one having open ear lobe; the other having a closed ear lobes. A single nucleotide polymorphism (SNP) is a type of gene variation that occurs most commonly in humans or homo sapiens. The SNP, rs769452, is a single difference in nucleotide that is associated with alzheimer's and subarachnoid hemorrhage in people. This change in nucleotide is classified as pathogenic meaning the diseases associated with it are caused by some virus or microorganism. However, fortunately very few people have developed major health complications as a result of a SNP. Researchers use them more as biological markers in order to prevent a likely disease from taking form should any genes associated with disease be found. Primer Design and Testing An Apolipoprotein E, APOE, is a type of gene that can influence the risk of becoming susceptible to the late-onset type of alzheimer's. There are three types of APOE gene called alleles that can greatly affect the chance of developing the disease. In other words by changing a nucleotide in a codon from a non diseased allele, CTG, then becomes a diseased allele containing the codon CCG. In our assignment we isolated the single nucleotide polymorphism and used its numerical position of 44907853 and designed a non diseased forward primer from one side of the DNA strand. We also had to create a non diseased reverse primer that was essentially the 220 base pairs away on other side of the DNA strand using the matching base pairs. Having the non diseased forward primer as well as the non diseased reverse primer, both were plugged into the genome.uscs.edu website and the result had successfully identified the original chromosome and location that were given at the beginning. Then the diseased codon primer and reverse primer were inserted with the result coming back with no matches due to the mutation not appear naturally in healthy humans. Non-diseased forward primer: AGCGGCCAGCGCTGGGAACT Non-diseased reverse primer: CAGGCCCCCCAAGACTTAGC Disease forward primer: AGCGGCCAGCGCTGGGAACC Disease reverse primer: CAGGCCCCCCAAGACTTAGC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||