BME100 f2016:Group9 W8AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
, and dNTP’s (http://www.promega.com/resources/protocols/product-information-sheets/g/gotaq-colorl ess-master-mix-m714-protocol/)
have the same forward primer and reverse primer
do, the samples will become cross-contaminated
PCR Reaction Sample List
Obtain a source of DNA wanted for PCR
Transfer the DNA into a special PCR tube to easily manipulate the temperature
Add Primer 1 to the special PCR tube because primers are very effective at copying DNA sequences
Add Primer 2 which attaches to second site
Add nucleotides to the tube to have a foundation to build more nucleotides off of
Add DNA polymerase which read the DNA and assign complementary nucleotides
Put the PCR tube into the thermocycler and start it OpenPCR program
INITIAL STEP: 95°C for 2 minutes NUMBER OF CYCLES: 25 DENATURE at 95°C for 30 seconds, Anneal at 57°C for 30 seconds Extend at 72°C for 30 seconds FINAL STEP: 72°C for 2 minutes FINAL HOLD: 4°C
Research and DevelopmentPCR - The Underlying Technology Once the DNA is inside the thermocycler, the thermocycler heats up the DNA to 95 degree Celsius which begins to unwind the DNA double helix into two separate strands.The thermocycler then drops to 50 degrees Celsius and at this temperature natural pairing of the DNA begins but since there are more primers than DNA strands, the primers lock on their target strands before they can naturally pair. The thermocycler then changes to 72 degrees Celsius and at this temperature the DNA polymerase is activated. The DNA polymerase then locates the primers and add the complementary base pairs. Soon the desired fragments will appear, primer one starts it and primer two ends it. The target sequence will soon become the majority. For example, after 30 cycles the process will have made over one billion target fragments. Table 1: What is the function of each component of a PCR reaction? Table 2: What happens to the components (listed above) during each step of thermal cycling? Table 3: DNA is made up of four types of molecules called nucleotides, designated as A, T, C and G.Base-pairing, driven by hydrogen bonding, allows base pairs to stick together. Which base anneals to each base listed below? Q4. During which two steps of thermal cycling does base-pairing occur? Explain your answers. The first step when base pairing occurs is when the temperature changes to 50 degrees Celsius, DNA natural pairs but since there are more primers the primers directly attach to the desired fragments and copies the DNA/nucelotides. DNA polymerase is added at 72 degrees Celsius which locates the primers and adds the complementary base pairs. A series of this process will produce a large amount of the desired DNA.
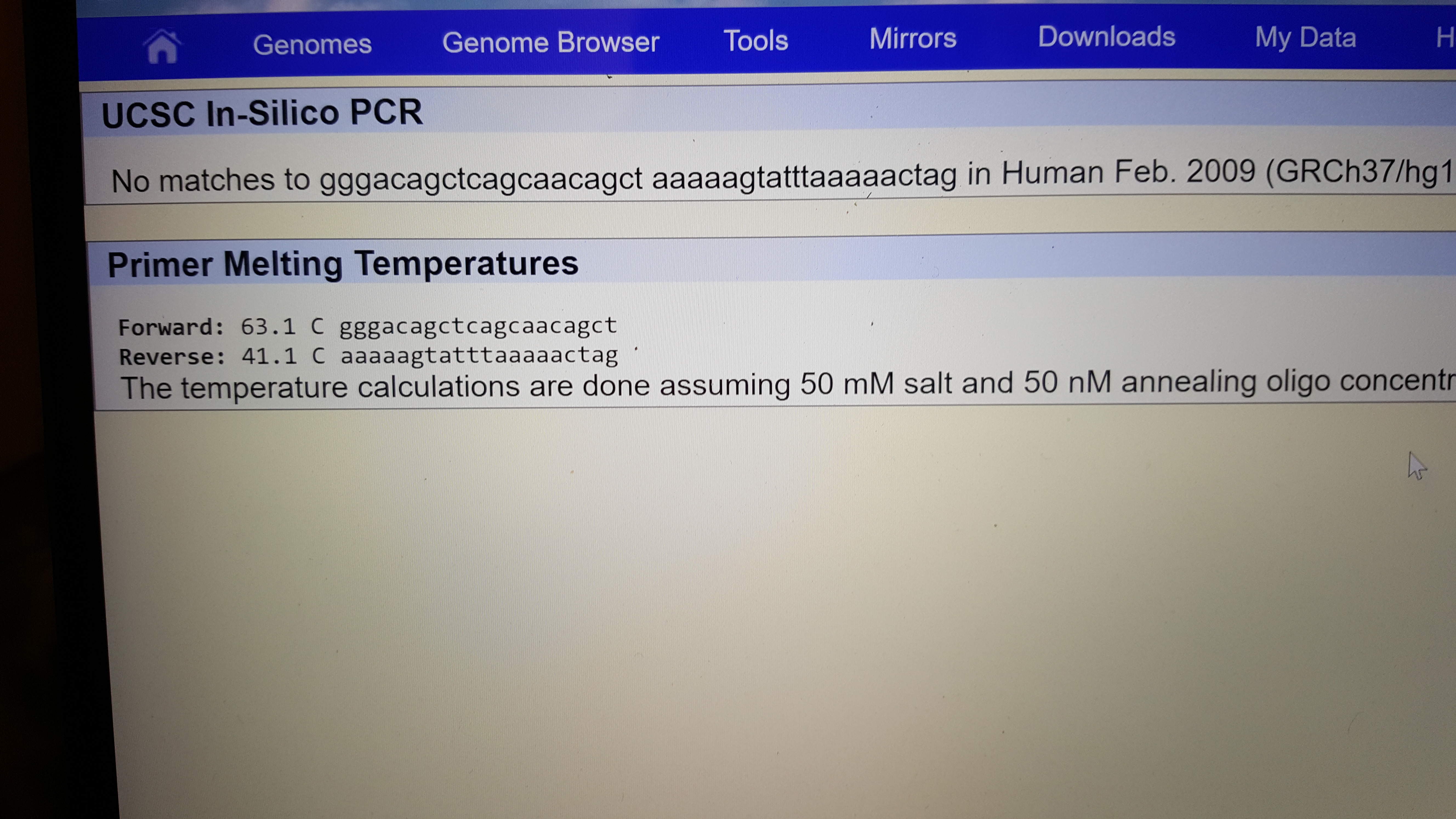
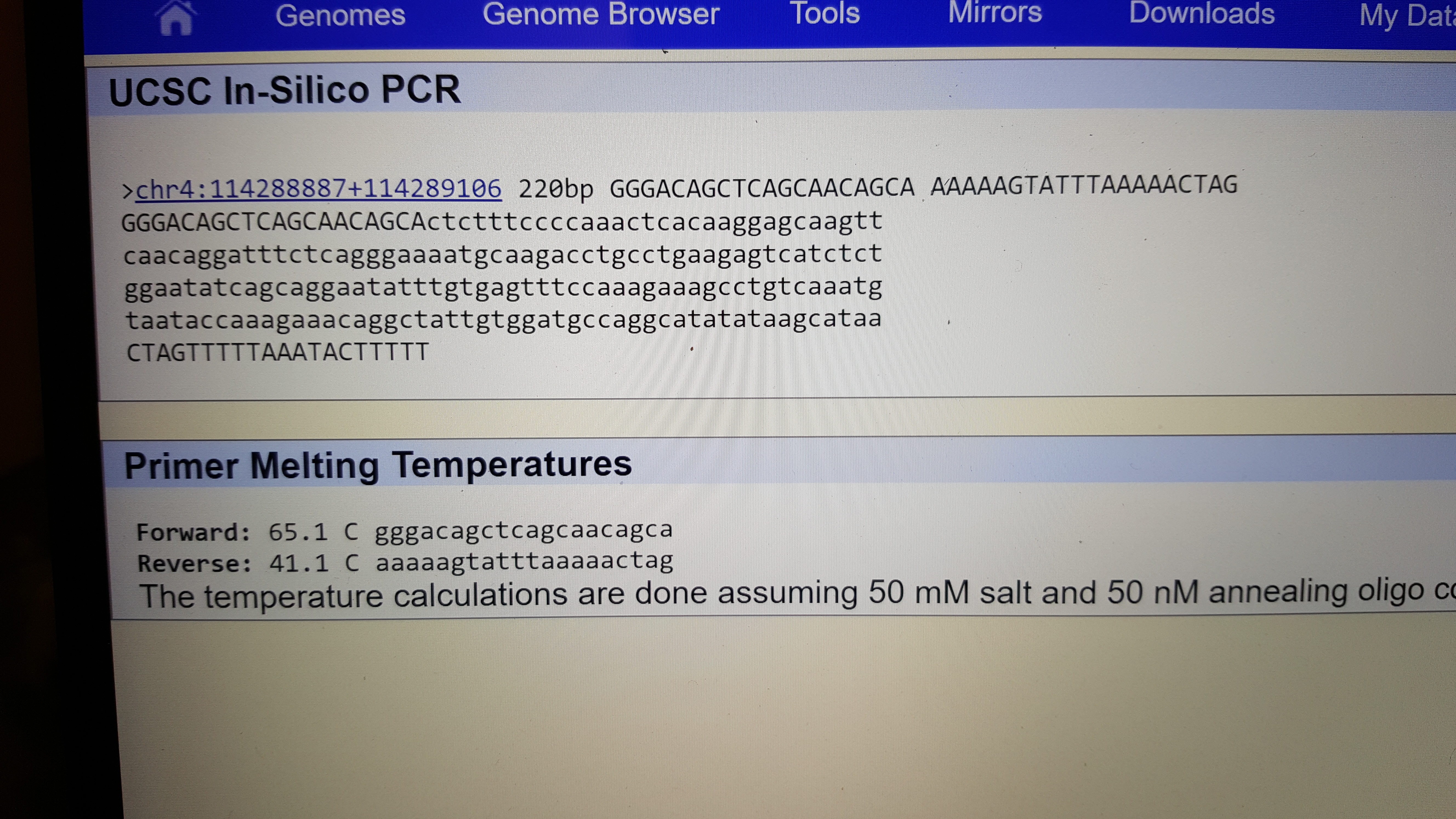
SNP Information & Primer DesignBackground: About the Disease SNP The disease SNP, single nucleotide polymorphism, is when there is a mutation or unexpected variation in a single nucleotide that happens in a specific portion of the genome. This is fairly rare happening at about 1 in every 1000 bases. This small difference can put an individual at a disadvantage because a mutation of a single base can increase the risk for Alzheimers. However, the presence of SNP provides a way to research the human genome by physical mapping which uses these variations as markers. The types of variations due to SNP can help predict how humans are effected by certain chemicals and how humans develop diseases. Nucleotide - Monomers of nucleic acid and contains a five carbon sugar, a phosphate group, and a nitrogenous base, these make up DNA. Polymorphism - A variation that develops different types of groups or individuals in a population. What species is this variation found in? Homo sapiens What chromosome is this variation located on? 4:113367751 What is listed as the Clinical Significance of this SNP? Pathogenic allele What condition is linked to this SNP? Cardiac Arrhythmia Syndrome What does ANK2 stand for? Ankyrin-2 What is the function of ANK2? ATPase binding enzyme binding cytoskeletal adaptor activity What is an allele? An alternative form of a gene located on a specific location on a chromosome. The disease associated allele contains what codon? ATC The numerical position is : 113367751 Non disease forward primer(20nt): GGG ACA GCT CAG CAA CAG CA The numerical position exactly 200 bases to the right of the disease SNP is: 113367951 Non disease reverse primer(20nt): AAA AAG TAT TTA AAA ACT AG Disease forward primer(20nt):GGG ACA GCT CAG CAA CAG CT Disease reverse primer(20nt): AAA AAG TAT TTA AAA ACT AG
Primer Design and Testing The results of the primer test revealed that the non disease primers indicated a 220bp sequence on the 4 chromosome.
| ||||||||||||||||||||||||||||||||||