BME100 f2016:Group5 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | ||||||||||||||||||||||||||||
|
OUR TEAMLAB 4 WRITE-UPProtocolMaterials
PCR Reaction Sample List
DNA Sample Set-up Procedure
OpenPCR program
Research and DevelopmentPCR - The Underlying Technology
The Function of Template DNA in a PCR reaction is to act as a road map for the whole reaction. This template DNA holds the desired gene, so the function of this strand is to hold the information of the desired gene and to allow a place for Taq Polymerase to replicate this gene.
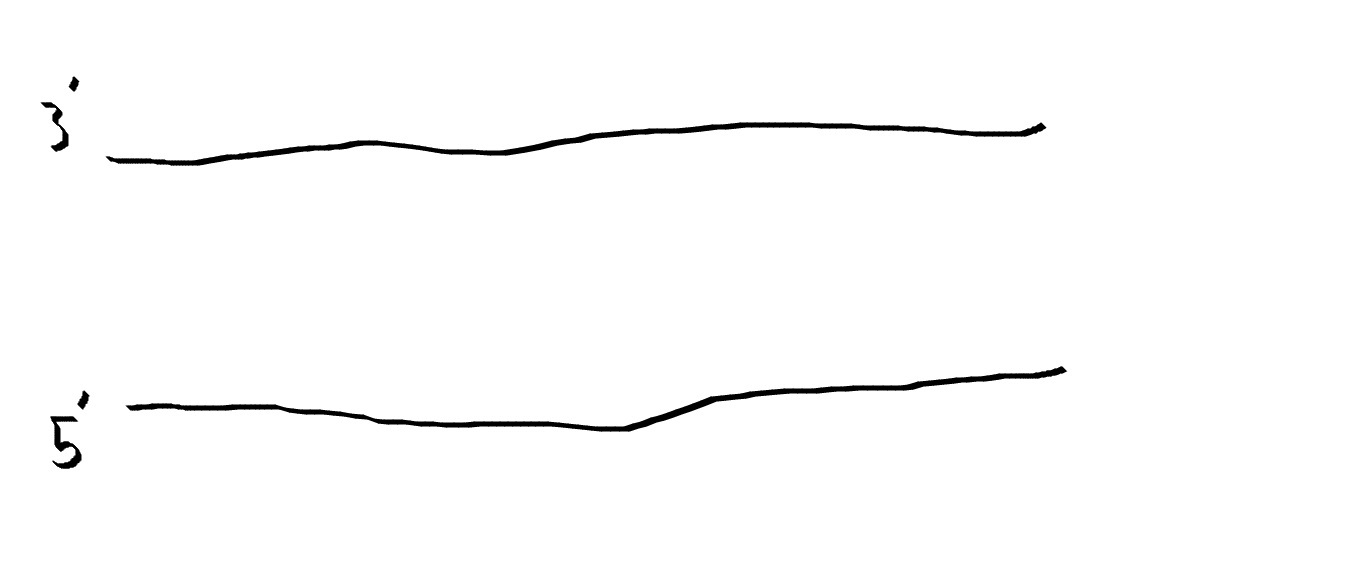
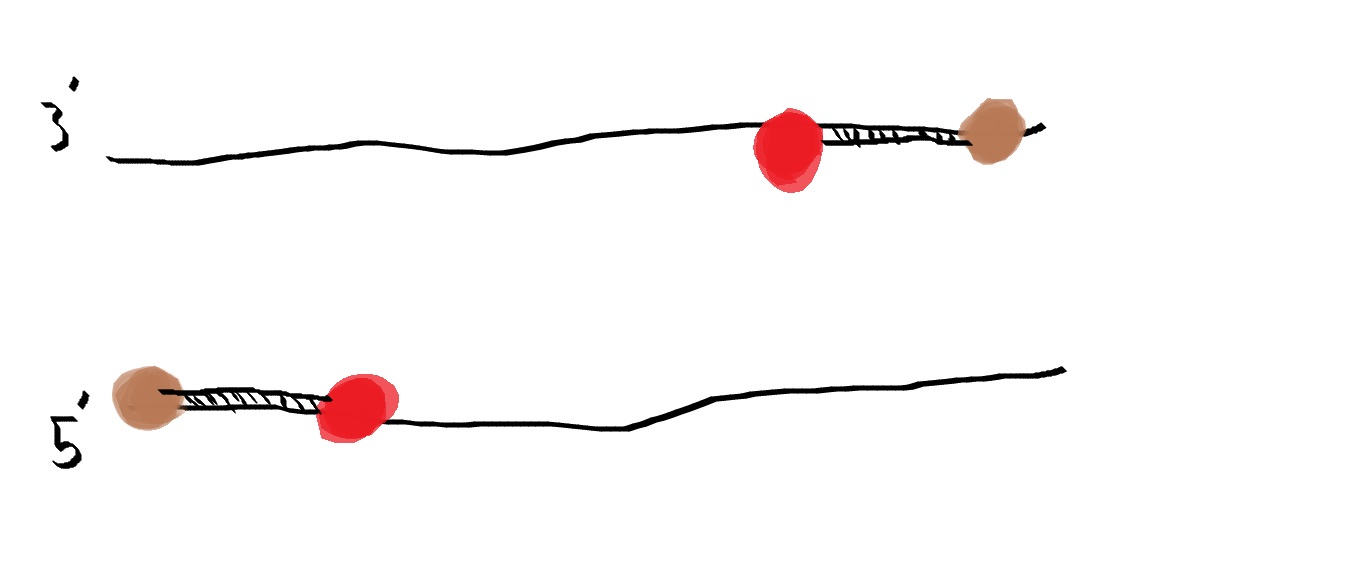
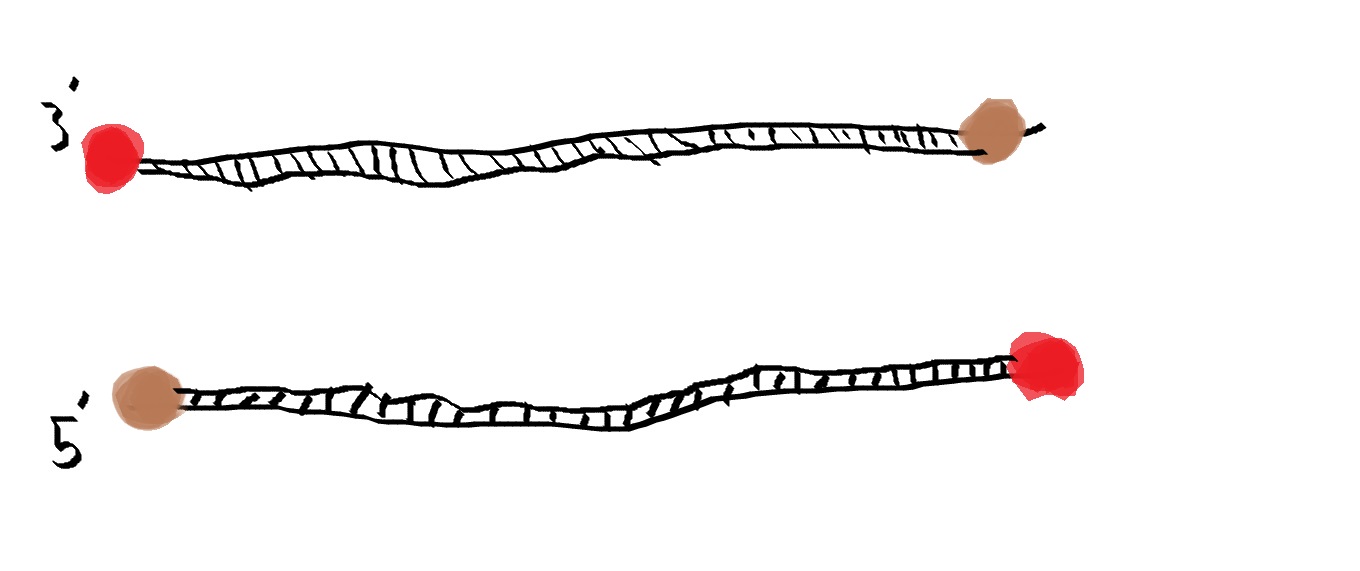
For a Polymerase Chain Reaction, two primers are needed in order for the reaction to take place. A 5' primer is need to bind to the 3' strand of DNA and vice versa. When these primers bind to the DNA, they act as a beacon for Taq Polymerase. They allow a site for Taq Polymerase to bind to DNA and tell Polymerase where to start replicating.
The function of Taq Polymerase is to zip across the single strand of DNA and to replicate a second stand of DNA as it goes along the first strand. It does this by reading the base pairs as it zips along and it pulls the counter part of that base pair from the environment, in this case the PCR tube, and binds it to the first strand of DNA. For example, if it comes across a T Nucleotide, it will pull an A from the environment and bind it to that T through hydrogen bonding.
A deoxyribonucleotide consists of 3 parts, a nitrogenous base, such as A, T, G, and C, a Deoxyribose sugar, and one phosphate group. DNA is fully comprised of deoxyribonucleotides bound together by the negative phosphates and the positive sugars binding together.The function of these is to act as building blocks for the new strand of DNA that Taq Polymerase creates. Witholut these, no new strand of DNA could be created, therefore PCR could not happen without the addition of these deoxyribonucleotides. What happens to each of the components during each step of thermal cycling? 95°C The Dna seperates the double helix and creates two single-stranded DNA molecules. 57°C The single strands of DNA try to pair up but there are more primer sequences than DNA strands in the tube and the primers go and lock onto their target before strands can rejoin. Primers will attach. 72°C Activates Taq polymerase and attaches to a single DNA strand, and adds complementary nucleotides onto strand. It continues until it gets to the end of strand and falls off. Stimulate Taq polymerase to copy strand. Which base pair anneals to who? The base pair Adenine bonds to Thymine using two hydrogen bonds while the base pair Cytosine binds to Guanine using three hydrogen bonds. During which two steps of thermal cycling does base-pairing occur? Base pairing occurs in the Final step and in the extend step of the thermal cycling. In the extend step of PCR, Taq Polymerase attaches to the DNA strand and it begins to create the second strand of DNA by pulling deoxyribonucleotides from the environment and adding them to the denatured template strand. Then in the final step, Taq Polymerase continues to work and continues to add base pairs to the strand of DNA for about 3 minutes. Since DNA is comprised of base pairs, these two steps are where base-pairing occurs within a Polymerase Chain Reaction. How Primers bind to DNA and how Taq Polymerase amplifies the DNA The primers are in brown while Taq polymerase is in red. DNA is denatured, meaning the double helix structure flattens out and then the two strands separate from one another, and now they are ready for the primers. The primers will attch to the sites they were designed to attach to and they will act as a landing site for Taq Polymerase. Polymerase attaches to the primers and from here it will start replication. Polymerase zips its way down the strand as it reads the base pairs and pulls lone Deoxyribonucleotides from the environment and bonds them to the strand it is reading. Once polymerase finished, the primers and polymerase will pull away from the DNA and wait for the next cycle of PCR to occur. Sources http://learn.genetics.utah.edu/content/labs/pcr/
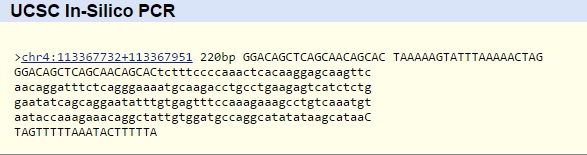
SNP Information & Primer DesignBackground: About the Disease SNP The disease SNP is on Chromosome 4, specifically at chromosome position 113367751 which can be found in humans. Because of the loss of ankyrin- B function it causes cardiac arrhythmia syndrome. The clinical significance listed of SNP is pathogenic. Its a single nucleotide variation and have A/C ancestral alleles. The gene ID for this is ANK2 (287) which stands for ankyrin 2. Ankyrin 2 is vital to cell motility, activation, and proliferation. Primer Design and Testing Non Disease Forward Primer: GGACAGCTCAGCAACAGCAC Non disease Reverse Primer: TAAAAAGTATTTAAAAACTA Diseased Forward Primer: GGACAGCTCAGCAACAGCAA Diseased Reverse Primer: TAAAAAGTATTTAAAAACTA Summary In this experiment, we took the human genome and found a certain SNP that we wanted to observe, and in order to observe it in abundance, we must perform PCR, or Polymerase Chain reaction. So, in order to make PCR work, we need primers to attach to the DNA strands so that Taq Polymerase can attach to the strands and replicate that certain SNP over and over again. So we made primers, one for the reverse strand and one for the forward strand for the non diseased allele DNA. The results that we obtained from testing these primers were that the primers worked successfully, we were able to prime the set of 220 base pairs that we need for the testing of the SNP. Next we made diseased primers which just included changing the last base pair of the forward strand, however these primers failed. The reason these failed was due to to the fact that the genome does not include the diseased allele, therefore the primer could not find a spot to attach to, therefore Taq Polymerase could not replicate that strand of DNA. Non Diseased Primers Diseased Primers |
||||||||||||||||||||||||||||