BME100 f2016:Group3 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4: Experimental DesignProtocolMaterials
First, collect necessary materials, as stated above. In order to fit all the samples inside the PCR machine you must cut the tubes in half, then label the sides of the tubes and place them into a rack. To start the lab, take the empty tube labeled “positive control” and insert 50 µL of the PCR reaction mix. In the same tube, transfer the positive control DNA (the primer mix) so that the tube holds 100 µL. Next, take the empty tube labeled “negative control” and repeat steps for “positive control” above with appropriate DNA (primer mix). Repeat steps used for “positive control” and “negative control” for patient 1 replicates 1, 2 and 3, and for patient 2 replicates 1, 2 and 3. Finally, place tubes in the heating block of a PCR machine.
OpenPCR program
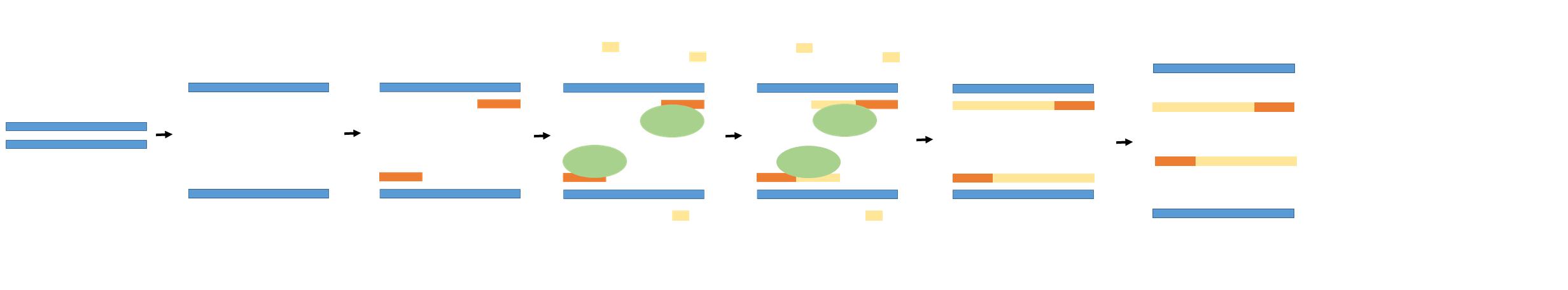
Research and DevelopmentPCR - The Underlying Technology PCR Functional Components (Q1) The components that drive a polymerase chain reaction are template DNA, primers, taq polymerase, and deoxyribonucleotides (dNTPs). The template DNA is the sample of DNA that contains the target sequence, or, the sequence being amplified by the PCR. The primers are short DNA sequences that attach to either end of the target sequence being copied - there are two primers that bound the target sequence. Taq polymerase is the enzyme that binds to the primer and attaches deoxyribonucleotides to extend the primer sequence PCR Steps (Q2) The initial step, when the tube is heated to 95°C for 3 minutes, is to ensure that the double-stranded DNA molecules are separated into two single stands for amplification. This is the denaturing of the sample happens next, at 95°C for 30 seconds, and is important to ensure that the double helix separates and creates two single-stranded DNA molecules. The anneal happens next at 57°C for 30 seconds, and the single-stranded molecules naturally try to pair up during this cycle, but the primer sequences bind with the DNA instead. It is extended next at 72°C for 30 seconds, and the DNA polymerase is activated and begins to attach the rest of the nucleotides onto the strand. The final step, 72°C for 3 minutes, is when the final copies are completed and all enzymes are removed from DNA. Finally, it is held at 4°C to stabilize the sample until it can be removed from the machine. Base Pairing During the Annealing and Extending Step (Q3, Q4) DNA nucleotide bases pair as follows, A (adenine) to T (thymine) and G (guanine) to C (cytosine). Base pairing is what allows the single stranded primer DNA to bind or anneal to the target DNA sequence during the annealing step and the taq polymerase then extends the primer DNA during the extending step by pairing bases as it moves down the single stranded target DNA. Base pairing enables the target DNA, through these two steps, to be amplified.
SNP Information & Primer DesignBackground: About the Disease SNP The disease SNP is a variation found in Homo Sapiens on the chromosome number 4. The listed clinical significance of the disease SNP is “with pathogenic allele”. An allele is defined as different forms of a gene that form due to mutation. The disease-associated allele of SNP is ATC, where the non-disease allele would be CTC. This disease SNP is linked to cardiac arrhythmia syndrome caused by loss of ankyrin-B function.
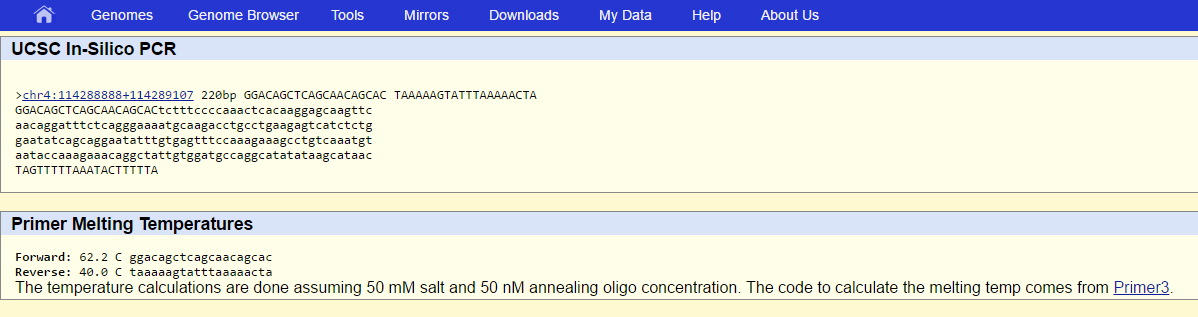
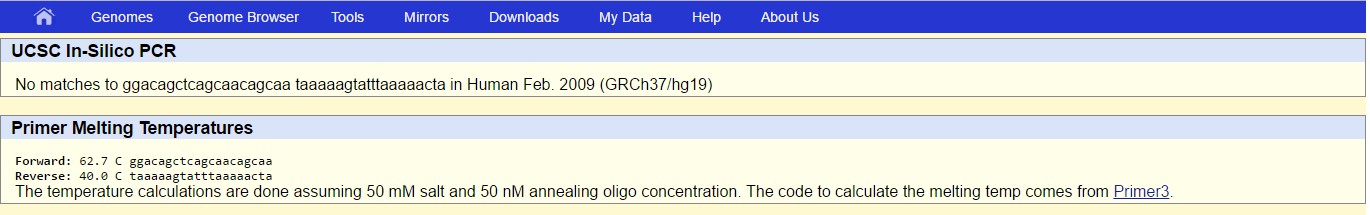
During the primer test we took the non-diseased forward primer from 5' to 3' and then changed the last base to make it diseased. We repeated this process with the reverse primer as well. We then plugged our results into the USC In-Sillico PCR website to find that our non-diseased primers resulted in an answer equal to the chromosome predicted, and our diseased primers had no results. |
|||||||||||||||||||||||||||||||||