BME100 f2016:Group1 W1030AM L4
| Home People Lab Write-Up 1 | Lab Write-Up 2 | Lab Write-Up 3 Lab Write-Up 4 | Lab Write-Up 5 | Lab Write-Up 6 Course Logistics For Instructors Photos Wiki Editing Help | |||||||||||||||||||||||||||||||||
|
OUR TEAM
LAB 4 WRITE-UPProtocolMaterials
(http://www.promega.com/resources/protocols/product-information-sheets/g/gotaq-colorl ess-master-mix-m714-protocol/)
have the same forward primer and reverse primer
do, the samples will become cross-contaminated
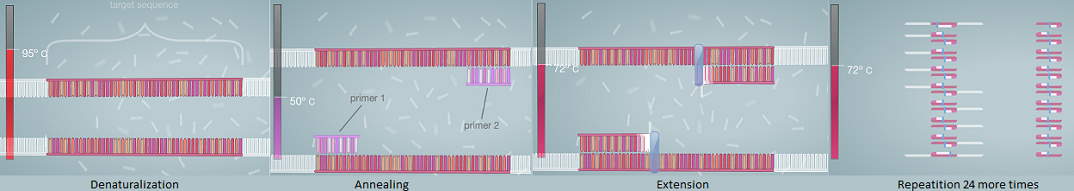
Step 1: Eight empty PCR tubes are each filled with a DNA-primer mixture with the help of a pipette. To avoid contamination, one pipette tip is used only once, then placed into waste cup. Step 2: The PCR mix and the other mix containing Taq DNA polymerase, MgCl2, and dNTP’s are carefully added into the PCR test tube. Step 3: PCR test tubes are placed into DNA thermal cycler.
OpenPCR program Having finished the set-up procedure described above, the properly prepared tubes are in the thermal cycler, the latter ready to be used. Step 4: Thermal cycler is set to 100°C. (thorough denaturalization) Step 5: The following cycle is done 25 times: for 30 seconds cycler is run on 95 °C (denaturalization), then cooled to 57°C for 30 seconds (annealing), then heated to 72°C for 30 seconds (extension). Step 6: Set thermal cycler to 72°C, run for 2 more minutes (thorough extension). Step 7: Finally cool content to 4°C (storage, conservation).
Research and DevelopmentPCR - The Underlying Technology The PCR Reaction and its components The Polymerase Chain Reaction, PCR, is used to analyze DNA. During this procedure, a specific segment of DNA is copied billions of times in a few hours. Four components of a PCR reaction are template DNA, primers, taq polymerase, and Deoxyribonucleotides (dNTP’s). The template DNA is the double-stranded DNA sample taken from the patient that will be copied. Artificial pieces of DNA, known as primers, attach to the DNA sample that is going to be copied. When two primers are used, one primer attaches to the bottom, and another to the top of the DNA piece, according to the rules of complementary base pairing. This stops the DNA polymerase enzyme from copying unwanted parts of the deoxyribonucleic acid. DNA polymerase enzymes are proteins that analyze the DNA code and attach corresponding nucleotides to create the DNA copies. In this process, taq polymerase is used because it is a special type of DNA polymerase that is able to work at higher temperatures in an aquatic environment. There are four types of Deoxyribonucleotides that are found in DNA code: Adenine, Cytosine, Guanine, and Thymine. These are the nucleotides the taq polymerase attaches to the strands of DNA. They are the foundation of DNA, which encode the genetic information. Thermal cycling In the initial step, the Thermal Cycler is set at 95°C for 3 minutes. Then the DNA is denatured at 95°C for thirty seconds. This is when the two matching strands of the double helix separate, becoming the two DNA templates. During the anneal stage at 57°C for 30 seconds, the primers connect to the matching parts of the templates. One at the top part of the DNA segment to be copied on one strand, one at the bottom of the segment on the other strand. The base Adenine (A) always anneals to Thymine (T), and Thymine always anneals to Adenine. Likewise, Cytosine (C) anneals to Guanine (G) and Guanine anneals to Cytosine. This is necessary because without the anneal, the DNA cannot elongate. When the DNA extends at 72°C for 30 seconds, the DNA polymerase enzyme is activated. It locates the primer attached to the DNA strand and begins building the copy with the addition of deoxyribonucleotides matching the template. This process is repeated several times. However, the denaturation (first) step needs only 30 seconds from the second cycle on. In every cycle, more and more copies are created because all the existing DNA become templates. In the final step, the Thermal Cycler is set to 72°C for 3 minutes while all the strands with a primer on it are copied. Then, it is held at 4°C to make sure all DNA strands form a double helix. This also prevents future contamination or degradation. Source of image: Learn.Genetics, University of Utah, Genetic Science Learning Center.
Base-pairing occurs during annealing and extension. First, the primers, these short, specific pieces of DNA connect to the part of the templates which are matching their base sequence. (For example, primer with sequence ACATGC would attach to the nucleotides of DNA in sequence TGTACG.) Then, with the help of the Taq polymerase enzyme, a new strand of DNA is created. For every nucleotide in DNA, a matching nucleotide floating in the solution is linked as the enzyme zips along the template.
SNP Information & Primer DesignBackground: About the Disease SNP This disease SNP, associated with rs35530544, deals with cardiac arrhythmia. SNP stands for single nucleotide polymorphism, which means that a single nucleotide is incorrectly changed, which leads to the disease. This particular one is found in the homo sapiens (humans), and is located on the fourth chromosome. Polymerase chain reaction is used to show if an individual suffers from this polymorphism, and the strategy below can be used to detect any disease SNP. Technology and medical science is progressing to the point where maybe this particular SNP can be targeted and changed, and as the "fixed" cells replicate, the disease could be eliminated from that individual, perhaps as early as in the womb.
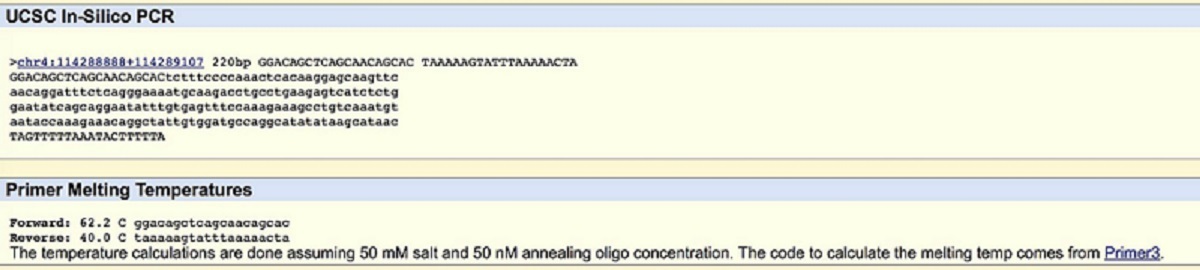
Part B Questions 1-15 A nucleotide is one of the components needed to create DNA and RNA structure. They are composed of a nitrogenous base, one sugar molecule, and one phosphoric acid. A polymorphism is a genetic variation that creates different types of the same species. It divides a species into distinct forms. It is found in the homo sapien species (human). The variation is located on the 4:113367751 chromosome. Its clinical significance is that it is pathogenic. SNP is linked to cardiac arrhythmia syndrome caused by loss of ankyrin-B function. ANK2 stands for ankryrin 2; three unique terms that were found were ATPase binding; cytoskeletal adaptor activity; enzyme binding. An allele is a form of a gene that results from a mutation. The disease-associated allele contains the ATC codon (the first C is changed to A). Its number position is 11336775. The non-disease forward primer is GGACAGCTCAGCAACAGCAC. Then 200 bases to the right is TAAAAAGTATTTAAAAACTA. The non-disease reverse primer GTGCTGTTGCTGAGCTGTCC. The disease forward primer is GGAAAGATAAGAAAAAGAAA. Lastly, the disease reverse primer TAAAAAGTATTTAAAAACTA.
| |||||||||||||||||||||||||||||||||